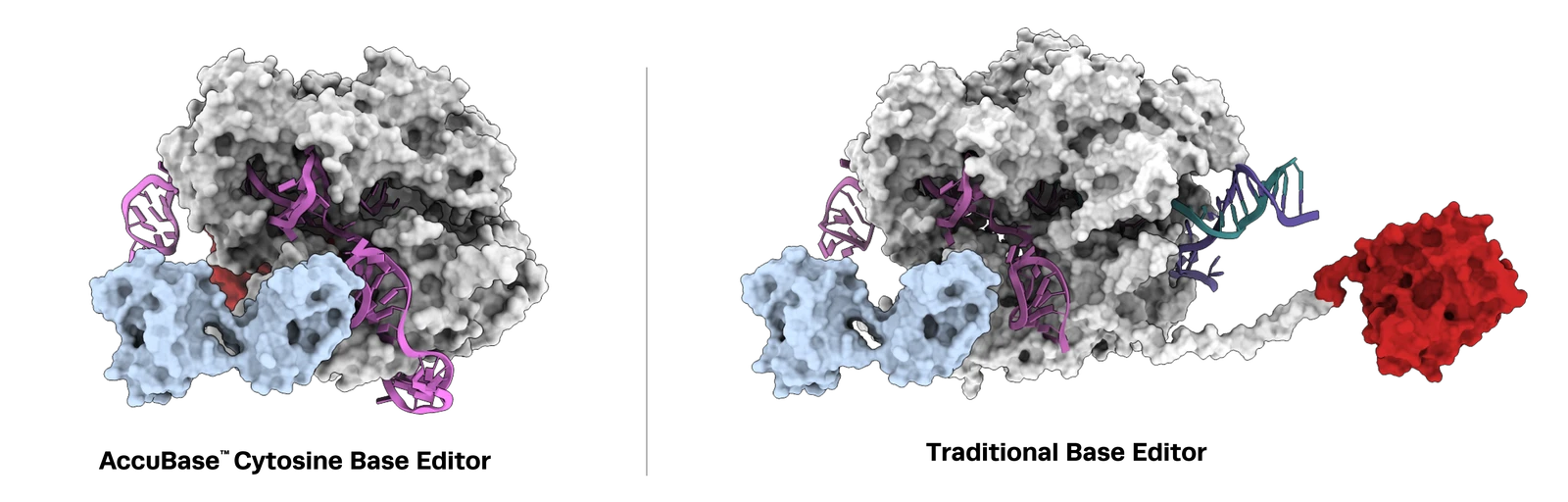
From RUO-to-GMP, Accubase, Synthego’s high fidelity engineered cytosine base editor (CBE) was purpose-built for high precision base editing in therapeutic applications. This engineered CBE contains a cytosine deaminase embedded within Cas9 nickase, that activates only when bound to the DNA target site, preventing unintended genome editing. Once bound to the DNA target site, Accubase exposes the deaminase enzyme enabling efficient and controlled conversion of cytosine (C) to thymine (T) within the 3-12 base editing window.
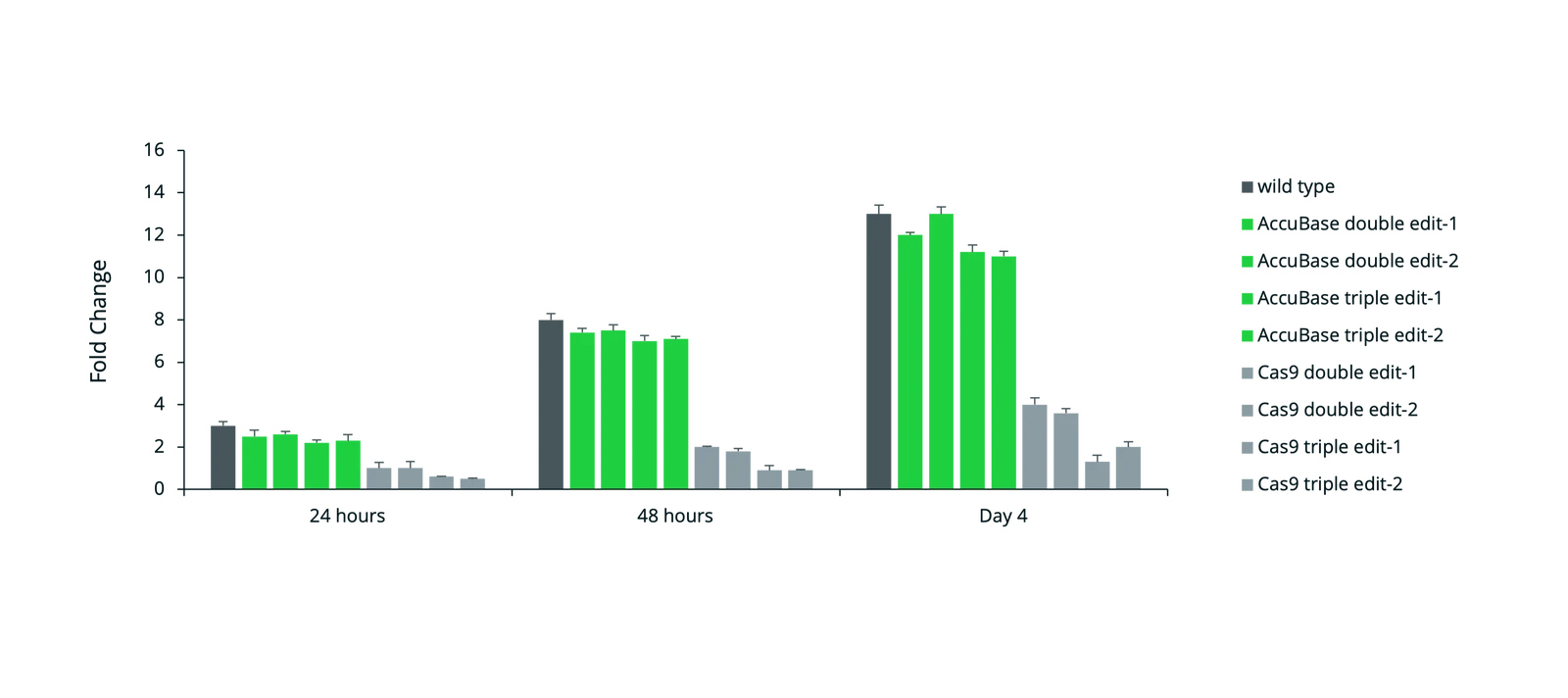
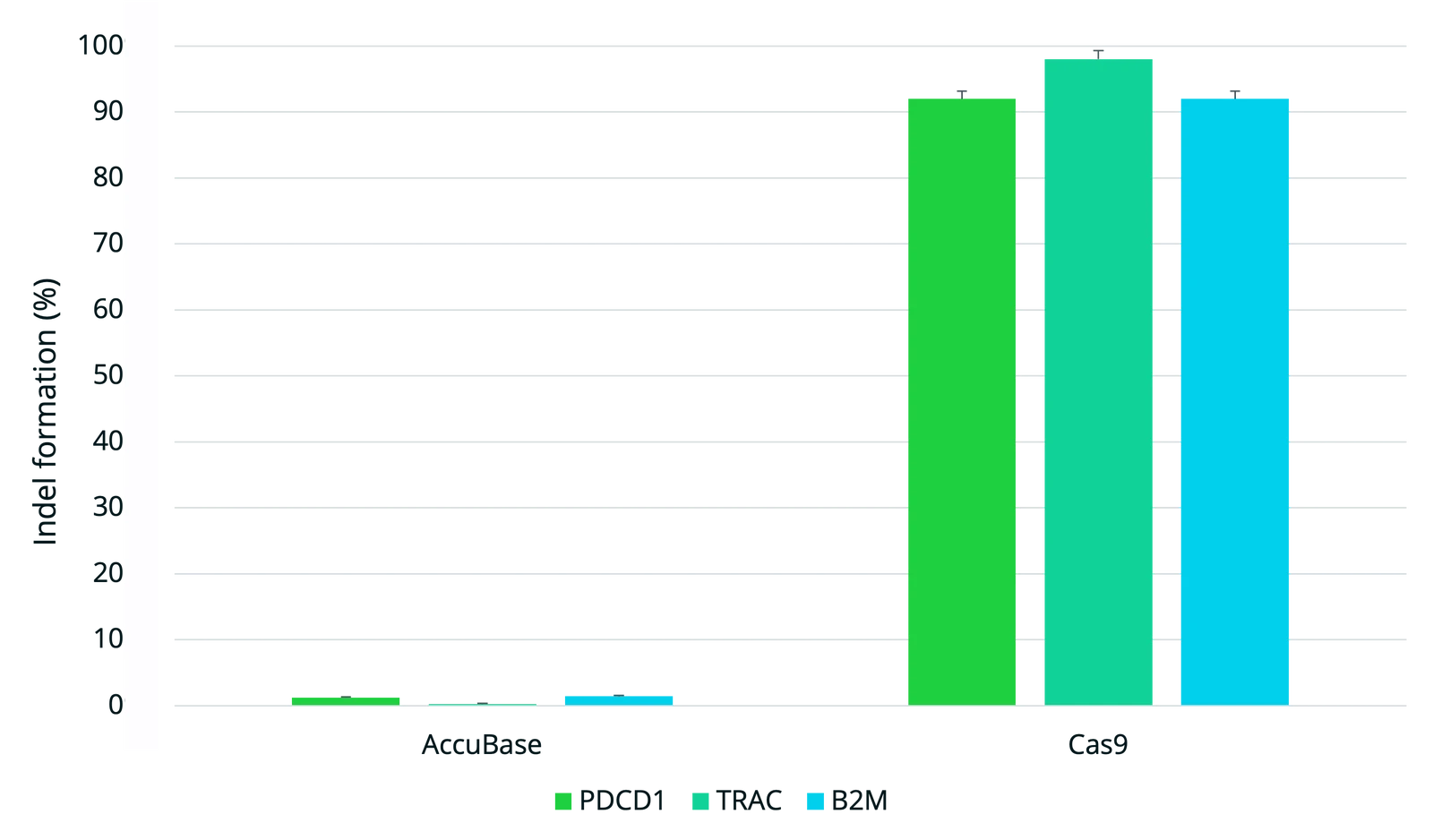
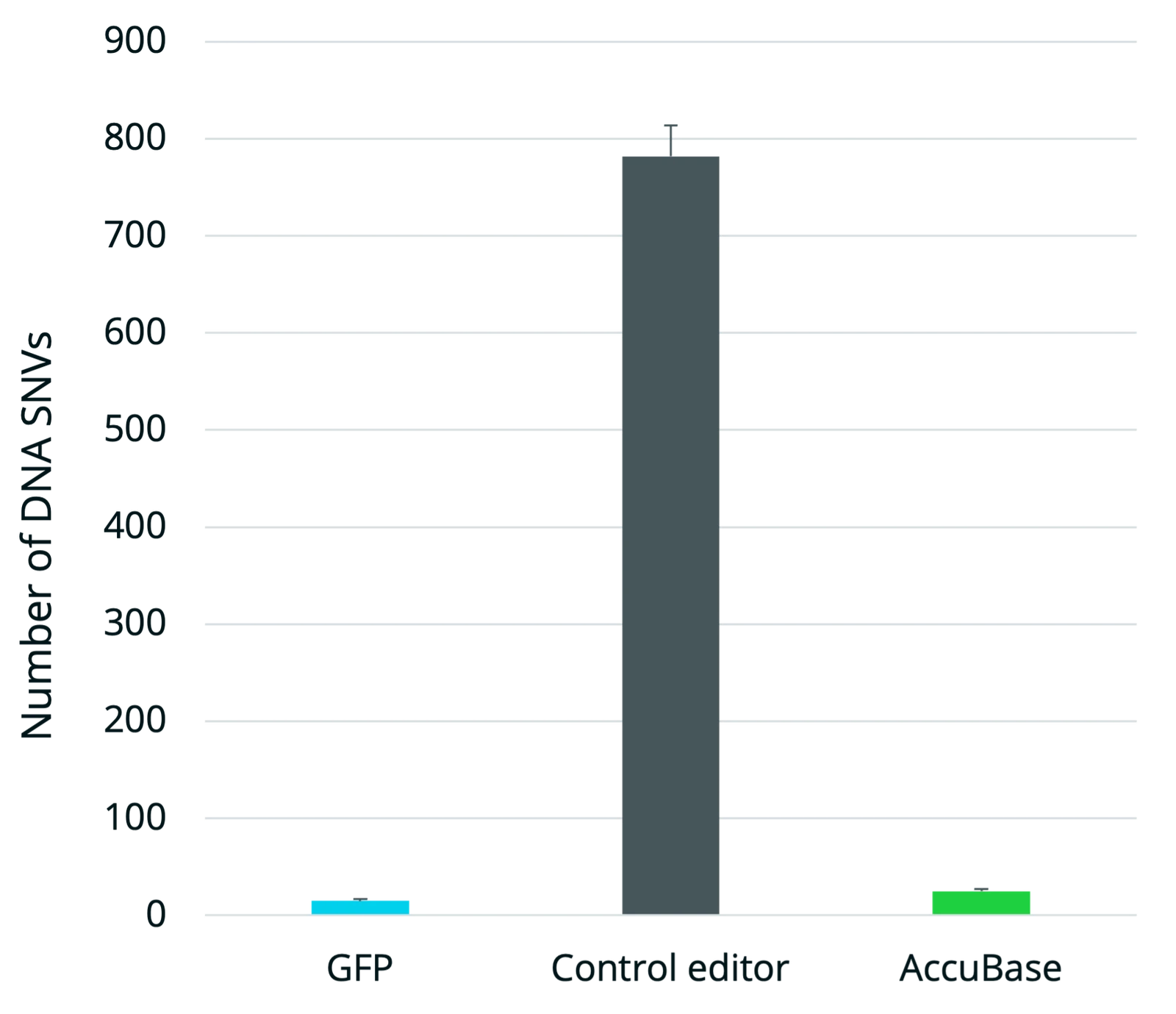
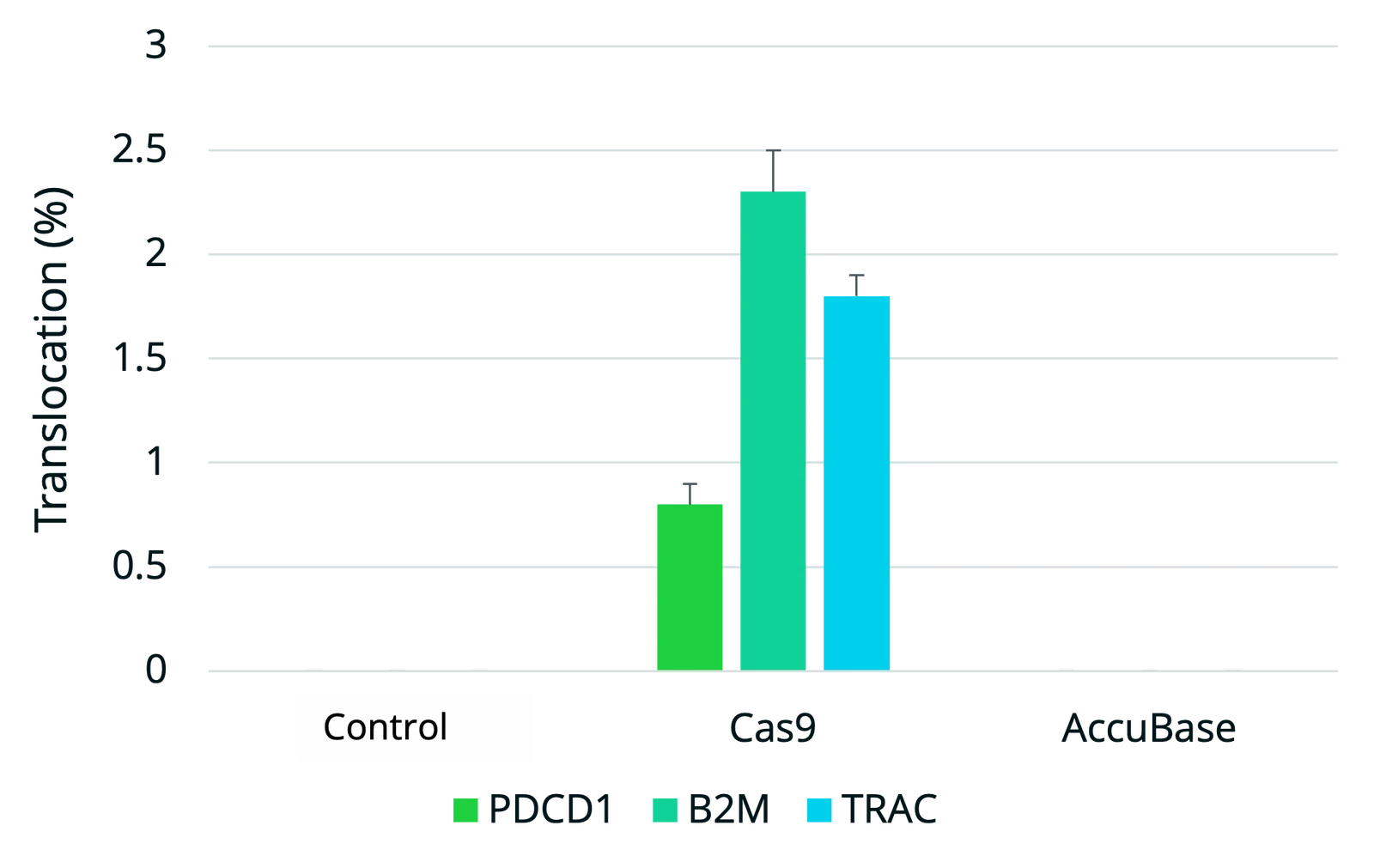
AccuBase CBE achieves high on-target editing in single and multiplexed applications with no detectable off-target editing or chromosomal translocations, offering an ideal solution for safe and effective base editing in cell and gene therapies. Where mRNA-delivered CBEs remain active in cells for longer increasing the chance of off-target editing, AccuBase is the only CBE supplied as a recombinant protein for transient expression. This allows scientists greater control over the duration and precision of editing, helping to minimize off-target risk potential and optimize outcomes in therapeutic genome engineering.
Using traditional cytosine base editors in cell and gene therapies presents key challenges. One major limitation is the use of mRNA delivery formats, which can result in prolonged protein expression within cells. This extended exposure increases the risk of off-target editing, leading to genotoxicity or unpredictable cellular behavior - unacceptable risks for cell and gene therapies. Furthermore, many current cytosine base editors exhibit off-target activity due to their continuous active state until they are eventually degraded in the cell. Therefore, finding a base editor that maintains high on-target precision without promiscuous off-target edits and translocation events has proven difficult - until AccuBase cytosine base editor.
AccuBase is also available in GMP. Bundling GMP AccuBase with Synthego’s best-in-class GMP gRNAs offers an advanced, comprehensive solution to streamline your therapeutic development. Both products are manufactured under true GMP standards, ensuring the highest level of quality, safety, and consistency from development to clinical use. This combination enhances specificity, reduces off-target activity, and ensures scalability, providing a robust base editing solution for IND-enabling studies and clinical-scale applications. Check out GMP AccuBase.