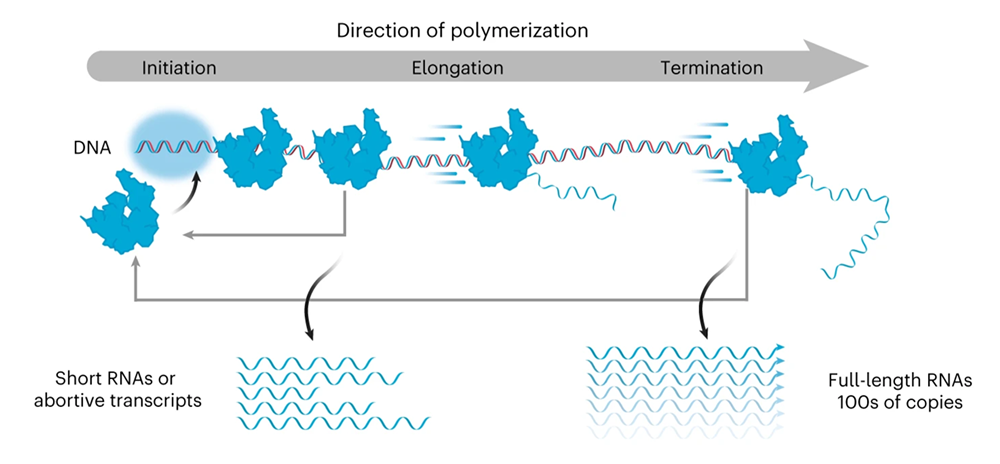
The bacteriophage T7 RNA polymerase is a polypeptide enzyme that is used to transcribe DNA to RNA. Unlike the majority of known RNA polymerase enzymes, T7 RNA polymerase consists of only one subunit. It is highly active and capable of synthesizing large amounts of full-length RNA transcripts in a relatively short time.
T7 RNA polymerase was discovered in the T7 bacteriophage and isolated from T7-infected E. coli cells in the late 1960s. It has since become a mainstay in molecular biology toolkits, being used to synthesize RNA for a variety of different applications.
The T7 RNA polymerase is one of the most commonly used because it has several key advantages over other RNA polymerases:
- Is a simple, single-subunit enzyme
- Recognizes its promoter sequence with very high specificity
- Produces high yield and high fidelity of RNA transcripts
- Works in short timeframes
- Terminates the process of transcription less often compared to, for example, E. coli RNA polymerase