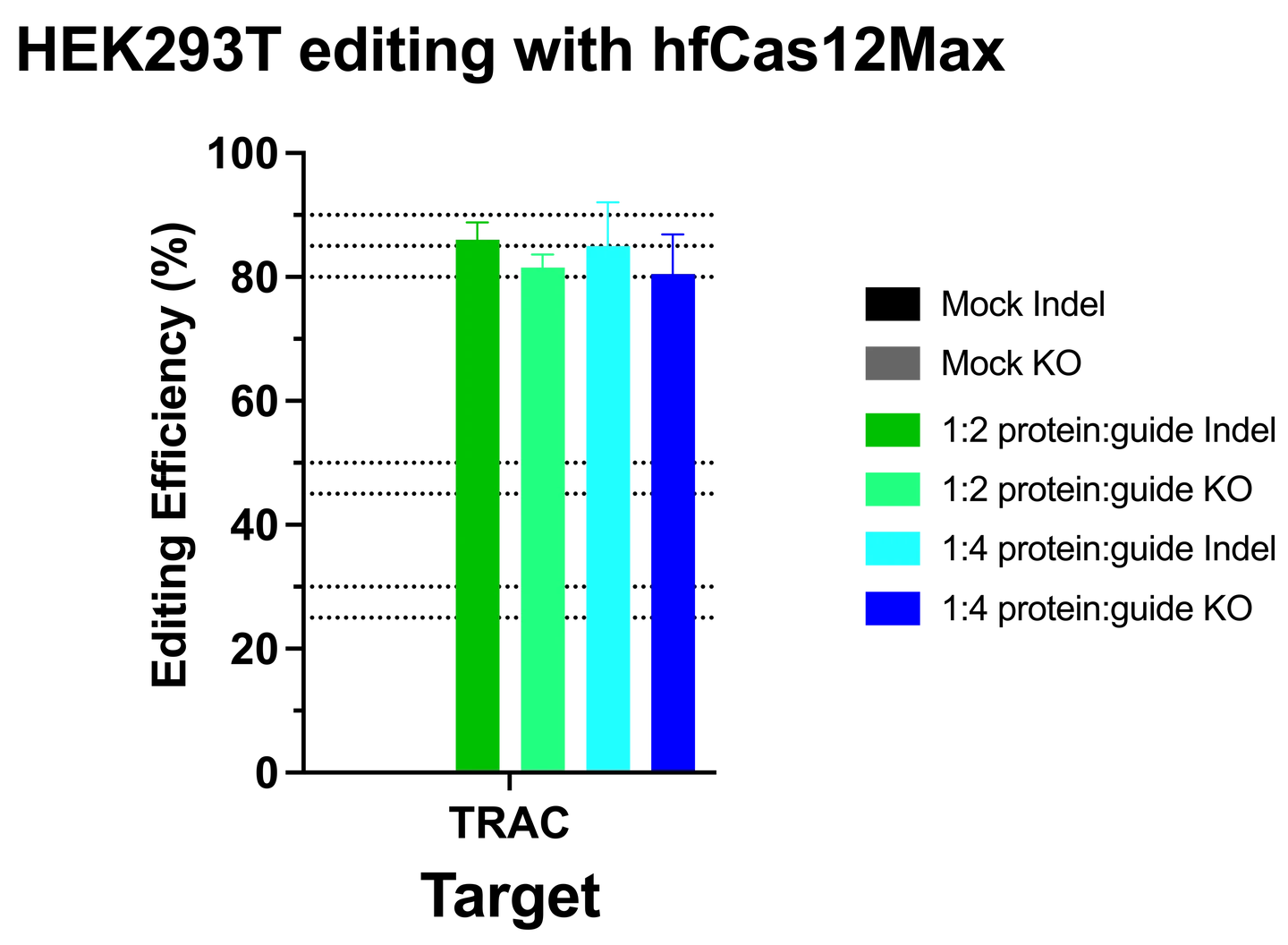
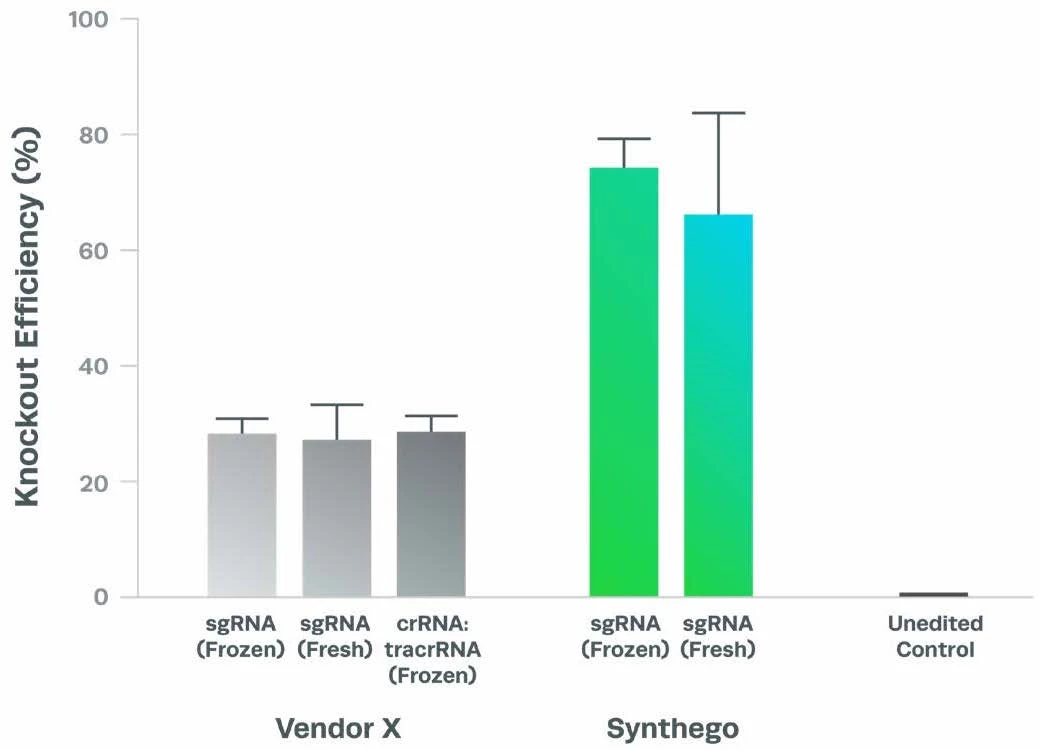
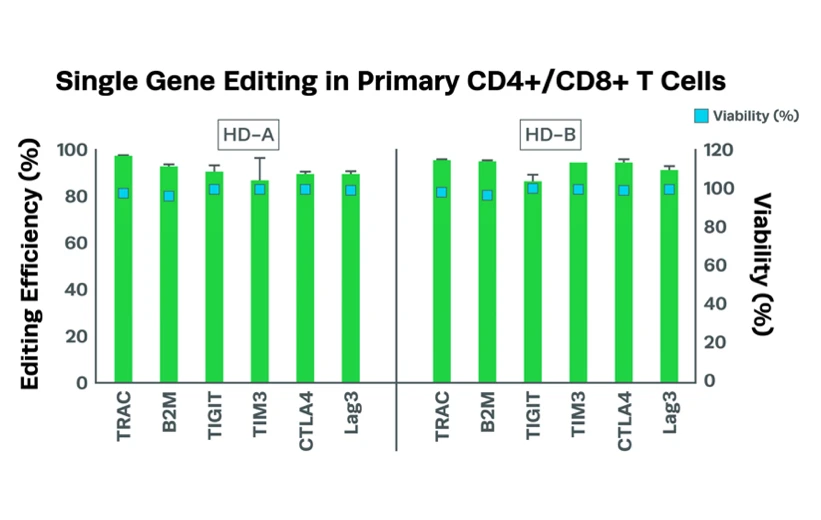
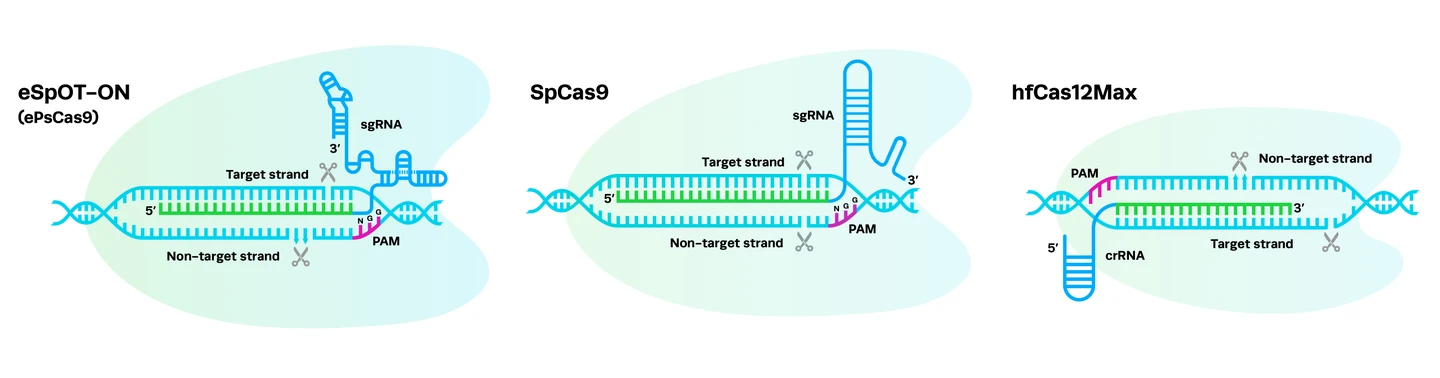
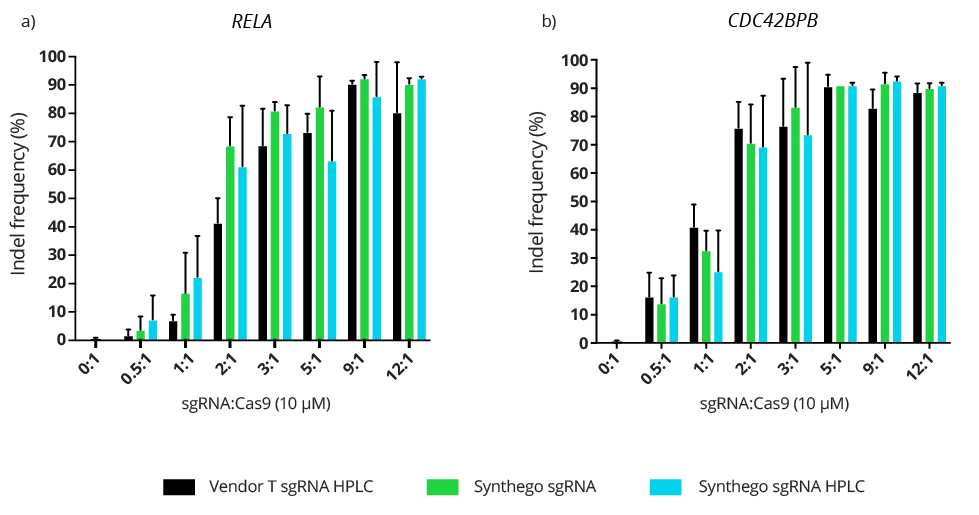
Complexing hfCas12Max nuclease with our best-in-class Research sgRNA achieves consistently high editing efficiencies. If you are seeking alternatives CRISPR gene editing methods, hfCas12Max is the nuclease you need for your next CRISPR experiment or therapeutic development.
Learn more
Webcast featuring eSpOT-ON
Explore More
eSpOT-ON Nuclease Protein Available Now
Explore More
Order eSpOT-ON Nuclease mRNA Now