The CRISPR field is moving fast! Don’t worry, we’ve got you covered. Check in every week for a quick summary of the biggest news and developments in genome engineering research so you can stay up to date with what’s happening in the world of CRISPR.
This week we report on a recently published study in which researchers from Harvard Medical School and the Wyss Institute for Biologically Inspired Engineering at Harvard University that used a CRISPR-based technique to study mammalian development.
The Basics of Developmental Biology
In sexually reproducing multicellular eukaryotes (i.e., you, me, your pet dog, the chicken you had for dinner last night), an organism begins as one cell. That single cell is referred to as a zygote: a combination of the male gamete (a sperm cell) and the female gamete (an egg cell) that have combined to produce a fertilized egg.
Zygotes are totipotent, which means that they have the potential to develop into any cell type, eventually organizing to form a complete organism. While scientists have learned a lot about the developmental process, the complete details of underlying mechanisms are still not fully understood.
We know that mammal development involves a complex web of linked processes that lead to cellular differentiation or specialization into individual cell types, tissues, and organs. It is thought that the path to creating all of the different cell types resembles a tree. At the base of the trunk is the zygote, with more specific cell types and structures developing as different branches as the tree grows.
Understanding Cellular Development and Differentiation
What isn’t so clear are the precise details of what that tree looks like and what seemingly tiny events happen at the roots of the tree impact the tip of the branches?
Understanding these disease-causing aberrations during the developmental process will not only influence how we identify, diagnose, and treat diseases, but they may also hold vital information about how to generate tissues and organs from stem cells to solve the current problems related to organ transplantation.
Using CRISPR to Generate Genetic Barcodes to Trace Cell Lineages
Over the years, scientists have used various methods to track an organism’s development. One common technique relies on labeling cells with dyes. This method has a significant limitation because the dye is diluted each time a cell divides, so approaches like these are not able to follow cells through multiple divisions and tracing development can only be done in short bursts.
In the past two years, CRISPR-Cas9 has emerged as a potent tool for uncovering the mechanism of this process in more detail than ever before.
Tracking cellular development from a zygote all the way to an adult animal necessitates two main conditions. First, there needs to be a reliable genetic marker that is transmitted every time a cell divides, and secondly, measurement of this marker must be easy.
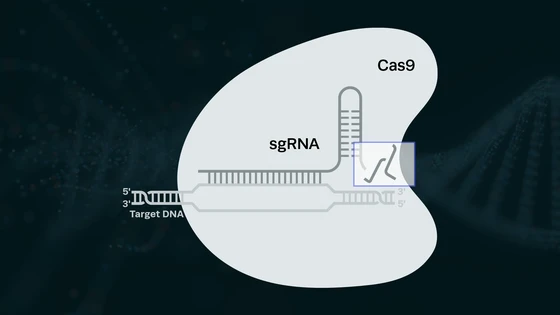
Introducing specifically designed genetic mutations into a predefined region in the genome can serve this purpose. In 2016, researchers developed a method to insert “genetic barcodes” into genomes as a marker to study cellular development. The technique involved first introducing short DNA fragments (termed “barcodes”) that would integrate into the genome at sites that do not disrupt any essential genes, and these barcodes serve as target sites for CRISPR-Cas9 machinery. CRISPR components could then be introduced into the cell at various stages of development, generating insertions and deletions of nucleotides in the inserted DNA sites.
The random nature of CRISPR indels created by this method allows individual cells to be tracked throughout the developmental process. For example, if one cell in an eight-cell embryo was edited to contain an insertion of two A bases at the barcode site, analysis of which cells in the adult that contain this same AA insertion would reveal all the cells that derived from that first edited cell.
The team tested this method in cell lines and in small animals such as zebrafish to trace back the cell’s lineage.
Tracking Cell Lineage in Developing Mice
Zebrafish are relatively small and simple organisms. To achieve similar results in a mammalian model, such as mice, needed several more target sites and subsequent mutations. The facts that the mouse gestation period is long and inside the female’s womb also complicated the process of introducing CRISPR machinery. In the recent study published in Science, Reza Kalhor and his colleagues at the Harvard Medical School found an interesting solution. They used homing RNAs (hgRNA) instead of standard CRISPR RNA guides in their experiments.
The concept of homing RNAs was introduced by part of the same team in a previous publication a couple of years ago. Consider the basic CRISPR mechanism in bacteria: the DNA sequences that encode guide RNAs are not cleaved by the Cas nuclease themselves because they do not contain PAM sequences. The researchers reverse-engineered this concept and developed homing RNAs, which target their own DNA sequence for cleavage, simply by adding in the PAM sequence in the guides. Their previous study has shown that homing RNAs editing results in far more diversity in accumulated mutations relative to conventional CRISPR guides.
Kalhor and his team bred a line of chimeric mice that contained 60 homing RNA barcoding sites scattered throughout their genomes. In theory, those 60 were enough to give a unique tag to each of an adult mouse’s 10 billion cells. Then they mated these mice with Cas9-expressing mice. This resulted in activated hgRNA in the progeny embryo that started editing their own DNA loci.
When the researchers sequenced DNA from different parts of the 12-day old embryo (primitive heart, limbs, egg yolk, and placenta), the shared mutation patterns enabled them to trace cellular histories of each part.
Further investigation of barcodes in the mouse embryos’ brain tissue demonstrated that the patterns were more similar between equivalent regions of the left and right side of the brain than between cells from different regions of the same side. This indicates that equivalent tissues on the left and right sides were formed from recent cell divisions. This pattern also suggests that the axis that runs from front to back of the brain forms before the one that runs from left to right; a timeline that neuroscientists have previously struggled to establish.
What’s Next For Genetic Barcodes and Developmental Biology?
“This method allows us to take the final developmental stage of a model organism and from there reconstruct a full lineage tree all the way back to its single-cell stage,” said co-author George Church, a Professor of Genetics at Harvard Medical School, in an interview with The Harvard Gazette. “It’s an ambitious goal that will certainly take many labs several years to realize, but this paper represents an important step in getting there.”
The researchers plan to refine and extend this new method to track the history of every single cell in a fully developed organism. They are also keen to explore new ways to more quickly read out barcodes from individual cells in the future.
“Being able to record cells continuously over time is a huge milestone in developmental biology that promises to exponentially increase our understanding of the process by which a single cell grows to form to an adult animal and, if applied to disease models, it could provide entirely new insights into how diseases, such as cancer, emerge,” said Donald Ingber, director of the Wyss Institute for Biologically Inspired Engineering at Harvard University, in an interview with The Harvard Gazette.
We’re not discussing humans scanning our own cells to see the most intimate family tree in the world, but this study is a significant step forward in our understanding of development from zygote to embryo. There is still a lot of work ahead for scientists to further develop and refine these findings, but we are excited to see what’s next!
More CRISPR in the News…
In Gene-Silencing Drug Approvals: Gene-silencing technology gets first drug approval after 20-year wait
For the first time, US regulators have approved a therapy based on RNA interference (RNAi). The medication, Alnylam’s patisiran, can now be used to treat a rare condition called hereditary transthyretin amyloidosis (hATTR). Patisiran prevents the production of a mutated version of the transthyretin protein, which is responsible for the overproduction of amyloids. When amyloid proteins build up, people can suffer peripheral nerve damage or polyneuropathy. "This approval is key for the RNAi field," James Cardia from RXi Pharmaceuticals in Massachusetts, which develops RNAi treatments, told Nature. "This is transformational." It is hoped that advances in RNA delivery will ultimately benefit researchers working with CRISPR, referencing the long wait time between technology innovation and approvals, Douglas Fambrough, chief executive of Dicerna, an RNAi-focused company in Cambridge, Massachusetts, said in a press release, “I hope they [CRISPR researchers] can do it more quickly than we did it.”
In Cas9 Ribonucleoprotein Complex Delivery: Targeted delivery of CRISPR-Cas9 ribonucleoprotein into arthropod ovaries for heritable germline gene editing
Investigators at Penn State College of Agricultural Sciences have developed a new method of using CRISPR gene editing technology. Currently, delivery of the Cas9 ribonucleoprotein (RNP) complex into embryos relies on microinjection – a challenging technique that is limited to a small number of species, and can be inefficient even in optimized organisms. The researchers developed a technology termed Receptor-Mediated Ovary Transduction of Cargo (ReMOT Control) to deliver Cas9 RNPs to the arthropod germline by injection into adult female mosquitoes. This innovative technique has been demonstrated to dramatically improve the efficiency of the method while eliminating the need for difficult microinjection of genetic material.
In CRISPR Startups: 10 High Potential CRISPR Startups To Watch Out In 2018
Explore Biotech released their list of top CRISPR companies, which covers companies using genome engineering to innovate in multiple different areas and applications. Synthego is proud to be included on the list, alongside many of the great companies we are supporting with our CRISPRevolution Kits and Engineered Cells products.
CRISPR 101 eBook
CRISPR has quickly become a standard laboratory tool for gene editing. As the adoption of CRISPR accelerates worldwide, up-to-date knowledge of the basics of CRISPR is essential for anyone in the field. From target identification studies to the recent breakthroughs in clinical trials, CRISPR is enabling scientists to unlock the power of the genome.
Download our CRISPR 101 eBook today to stay up to date on all your CRISPR basics and get the best results in your CRISPR experiments!

Cell Engineering 101
CRISPR has ignited a revolution. Although it’s a relatively recent discovery in the history of biotechnology, CRISPR has quickly become a standard laboratory tool and cell engineering is transforming research. One of the most widely used applications of CRISPR is knocking out specific genes in cell lines to interrogate gene function.