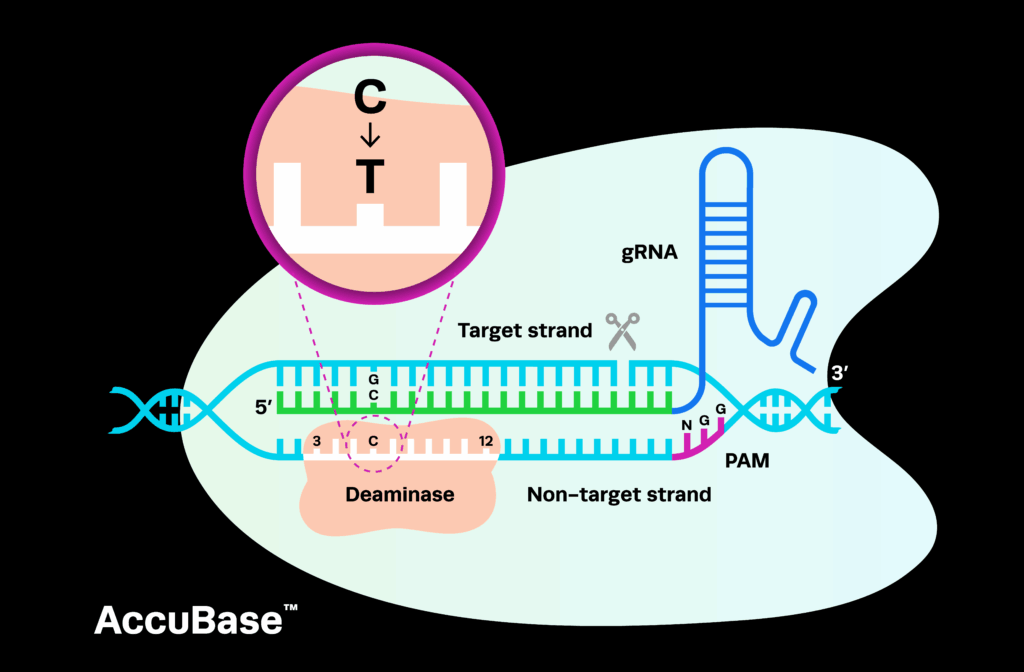
Synthego’s CRISPRevolution synthetic RNA kits are now being used by scientists throughout the world to achieve unparalleled CRISPR genome editing with minimal time and labor at a fantastic price point. We decided to find out what researchers thought about using our synthetic sgRNA kits, so we spoke with scientist Professor Joseph Miano from the University of Rochester to discuss his experiences.

Kevin Holden: Hi Joe! Could you tell us a little bit about your research?Dr. Joseph Miano: My research is focused primarily on understanding how genes are turned on and off. We make subtle substitutions in the regulatory elements involved in controlling gene expression in mice. Then, we study the genes that are under control of those regulatory elements to determine how important they are for function. The end game of a lot of our work is to interrogate elements for variants in human populations, and those variants, often times, are associated with diseases.
Our ultimate goal is to model single nucleotide polymorphisms in non-coding sequences in mice to gain more insight into the genetic basis of some diseases.
Dr. Joseph Miano: My research is focused primarily on understanding how genes are turned on and off. We make subtle substitutions in the regulatory elements involved in controlling gene expression in mice. Then, we study the genes that are under control of those regulatory elements to determine how important they are for function. The end game of a lot of our work is to interrogate elements for variants in human populations, and those variants, often times, are associated with diseases.
Our ultimate goal is to model single nucleotide polymorphisms in non-coding sequences in mice to gain more insight into the genetic basis of some diseases.
KH: What specific diseases are you focused on?JM: I am a vascular biologist by training, but in the recent years, primarily due to CRISPR technology, I have become more open minded towards organ systems. While most of the work therein is related to blood vessels and diseases, such as atherosclerosis or transplant arteriopathies, in the recent years, we have also ventured into kidney disease and GI diseases. Basically, wherever the CRISPR phenotype takes us, we follow that trail; if it’s an off blood vessel trail, that’s fine with us.
JM: I am a vascular biologist by training, but in the recent years, primarily due to CRISPR technology, I have become more open minded towards organ systems. While most of the work therein is related to blood vessels and diseases, such as atherosclerosis or transplant arteriopathies, in the recent years, we have also ventured into kidney disease and GI diseases. Basically, wherever the CRISPR phenotype takes us, we follow that trail; if it’s an off blood vessel trail, that’s fine with us.
KH: How long have you been using CRISPR for your research?JM: I was first introduced to CRISPR at the Cold Spring Harbor Laboratory in August, 2012 by Jennifer Doudna, who gave a keynote lecture. I was blown away! Her paper came out shortly thereafter, and then the avalanche began in early 2013. When I saw all that work coming out in early 2013, that was the turning point. So, it’s probably been five years now.
JM: I was first introduced to CRISPR at the Cold Spring Harbor Laboratory in August, 2012 by Jennifer Doudna, who gave a keynote lecture. I was blown away! Her paper came out shortly thereafter, and then the avalanche began in early 2013. When I saw all that work coming out in early 2013, that was the turning point. So, it’s probably been five years now.
KH: When you first started using CRISPR, do you remember exactly how you were doing it, how you would generate your guides and the nuclease?JM: Yes, very arduously. The very first experiments involved introducing plasmids in the mouse, and they were an abysmal failure! In those early years (2013 and 2014), we were all sort of in the dark, trying to figure out the best way to do this. We finally found our way to the synthetic RNA approach, and then RNPs came along. Now, we totally use RNPs!
JM: Yes, very arduously. The very first experiments involved introducing plasmids in the mouse, and they were an abysmal failure! In those early years (2013 and 2014), we were all sort of in the dark, trying to figure out the best way to do this. We finally found our way to the synthetic RNA approach, and then RNPs came along. Now, we totally use RNPs!
KH: How did you first hear about Synthego?JM: Probably through a Google CRISPR discussion group. Again, in the early years there were all kinds of questions: what kind of guide should we use? What are the ideal concentrations? How do we genotype? When I saw Synthego, I thought, “Well, that’s a pretty cool name.” Then I think we met in person at a meeting shortly thereafter, about a year and half ago or so. You were very kind and responsive.
JM: Probably through a Google CRISPR discussion group. Again, in the early years there were all kinds of questions: what kind of guide should we use? What are the ideal concentrations? How do we genotype? When I saw Synthego, I thought, “Well, that’s a pretty cool name.” Then I think we met in person at a meeting shortly thereafter, about a year and half ago or so. You were very kind and responsive.
KH: Did you use CRISPR before you came to your current research institute at the University of Rochester?JM: I didn’t even know what CRISPR was other than Doudna’s keynote lecture prior to 2013/2014. I was at the University of Rochester at the time, performing knockouts the old fashioned way. That’s where that started.
JM: I didn’t even know what CRISPR was other than Doudna’s keynote lecture prior to 2013/2014. I was at the University of Rochester at the time, performing knockouts the old fashioned way. That’s where that started.
KH: Did you come to University of Rochester right after finishing your PhD?JM: No, I went from Eric Olson’s lab at the University of Texas MD Anderson Cancer Center to the Medical College of Wisconsin for, what I call, my “super postdoc”, and after three years came to the University of Rochester, where I have been ever since.
JM: No, I went from Eric Olson’s lab at the University of Texas MD Anderson Cancer Center to the Medical College of Wisconsin for, what I call, my “super postdoc”, and after three years came to the University of Rochester, where I have been ever since.
KH: Can you tell us a little bit about the University of Rochester, about the facilities and the type of research?JM: The University of Rochester is noted mostly for its neurology and immunology related work, but less so for cardiovascular related work. It was an attractive option for me to move there because they were starting up a new Cardiovascular Research Institute. Also, Rochester is my hometown; it was an easy decision to go back home.
JM: The University of Rochester is noted mostly for its neurology and immunology related work, but less so for cardiovascular related work. It was an attractive option for me to move there because they were starting up a new Cardiovascular Research Institute. Also, Rochester is my hometown; it was an easy decision to go back home.
KH: How has using synthetic sgRNAs and the RNP format been beneficial to your genome engineering workflow?JM: It’s been a huge benefit because now my postdocs are free to do other research. They don’t have to synthesize the guide RNA, purify it, ship it off to be analyzed, and wait for all that to be done. That process took, on an average, two weeks from conception to actual mice injection. Now, we just order the sgRNA, wait for a week, then simply digest DNA in a tube, which takes about 30 minutes. Availability of these easy options has revolutionized the work in our lab; we can do more now, and quicker.
JM: It’s been a huge benefit because now my postdocs are free to do other research. They don’t have to synthesize the guide RNA, purify it, ship it off to be analyzed, and wait for all that to be done. That process took, on an average, two weeks from conception to actual mice injection. Now, we just order the sgRNA, wait for a week, then simply digest DNA in a tube, which takes about 30 minutes. Availability of these easy options has revolutionized the work in our lab; we can do more now, and quicker.
KH: The digest that you mentioned — is that an in vitro test on a PCR product to make sure the guides will cut?JM: Yes, and in every case thus far, we have had a one-to-one correspondence. What cuts in a tube, works in the mouse. I have heard from various people that isn’t always the case, but in our experience it’s rare for the guide RNA to work in vitro and not in the mouse.
JM: Yes, and in every case thus far, we have had a one-to-one correspondence. What cuts in a tube, works in the mouse. I have heard from various people that isn’t always the case, but in our experience it’s rare for the guide RNA to work in vitro and not in the mouse.
KH: A follow up question to the changes in workflow using RNPs: what is the difference in the time required for generation of a knockout mouse from the pre-CRISPR days to today with CRISPR using synthetic sgRNA and RNPs?JM: In the pre-CRISPR days, on average, it would be 18 months to forever (in those cases where we could never get germline transmission or even get ESL targeting using the old way). Now, I think we have a record of two and half months from conception to generation of a genotype-positive founder mouse. That’s pretty remarkable!
In the old days, there were projects where it would take longer than that just to make the targeting factor. It’s a huge game changer.
JM: In the pre-CRISPR days, on average, it would be 18 months to forever (in those cases where we could never get germline transmission or even get ESL targeting using the old way). Now, I think we have a record of two and half months from conception to generation of a genotype-positive founder mouse. That’s pretty remarkable!
In the old days, there were projects where it would take longer than that just to make the targeting factor. It’s a huge game changer.
KH: Lastly, what is your favorite thing about Synthego?JM: I like the codes, the freebies. I love the coasters. I love the t-shirts; the t-shirts rock! The website is really shaping up to be nice. Oh yeah, you and Jessica Roginsky are pretty cool too!
JM: I like the codes, the freebies. I love the coasters. I love the t-shirts; the t-shirts rock! The website is really shaping up to be nice. Oh yeah, you and Jessica Roginsky are pretty cool too!