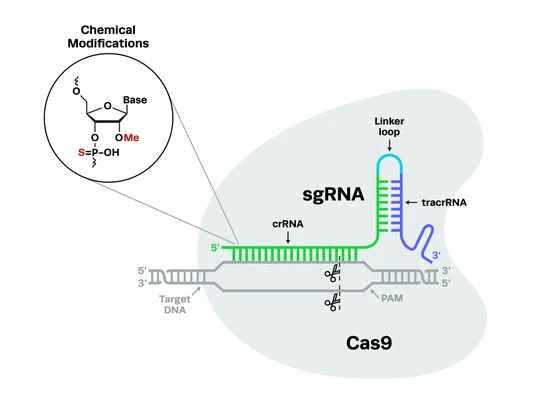
he field of genome engineering is recently abuzz with CRISPR updates. The CRISPR technology is a two-component system that allows precise recognition and editing of genomic sequences. Researchers are harnessing its potential of editing genetic mutations for developing curative therapies, but CRISPR’s potential in diagnostics is only recently being explored.
Now, Kiana Aran, a professor at Keck Graduate Institute at Claremont, California, and her team have developed CRISPR-CHIP, an electronic biosensor that uses CRISPR to detect specific genes in genomic DNA. Their findings have been published in Nature Biomedical Engineering today.
The Need for CRISPR-CHIP
The CRISPR system involves two components: the guide RNA is complementary to the target sequence and recognizes it, and the Cas nuclease cuts at this site. While most researchers focus on the editing function of CRISPR, Dr. Aran wanted to explore alternative directions.
"Everyone is thinking about the therapeutic application of CRISPR, but sometimes we forget that the first thing that CRISPR does is searching,” says Dr. Aran. "
Traditional diagnostic methods for nucleic acid detection depend on heavy instrumentation and time-consuming assays. Recently, CRISPR has been used for diagnostic kits—SHERLOCK, HOLMES, and Mammoth Biosciences’ kits being prime examples. However, their approach of using Cas13a and Cas12a nucleases for diagnostic detection requires amplification of signal for a clear readout.
Aran wondered if she could devise an even simpler gene-detection system that requires minimal reagents, effort, and time.
A Graphene-Based Electronic Platform Combined With CRISPR: How It Works
Dr. Aran used her electrical engineering background and biomedical engineering expertise to solve this problem. She combined the powerful search potential of CRISPR with a graphene-based field effect transistor (gFET) platform for sensitive readout to develop CRISPR-Chip.
The principle of CRISPR-Chip is simple. The gFET platform comprises a graphene layer that connects source and the drain electrodes. Aran and her team immobilized ribonucleoproteins (RNPs)—complexes of dead Cas9 protein conjugated with single guide RNAs—on the graphene layer. Dead Cas9 (dCas9) is catalytically inactive, i.e. it recognizes but does not cut the target DNA. The graphene platform is sensitive to the adsorption and interaction of charged molecules at its surface. Therefore, when a voltage is applied across the surface, binding of target DNA to the RNPs translates to a change in electrical current, which makes for a simple readout.
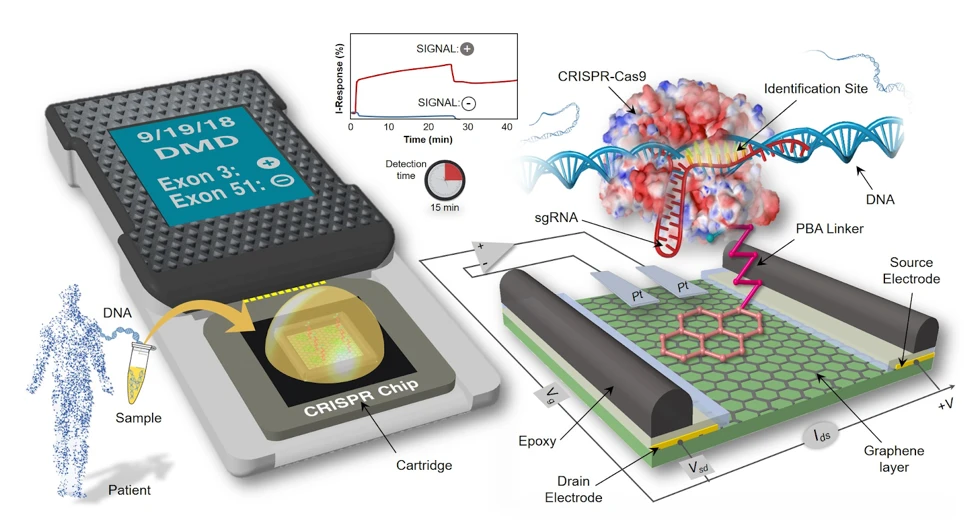
CRISPR-Chip Detects Target Genes in Genomic DNA
The researchers first tested their system on PCR amplicons, which are less complex to deal with than genomic DNA. RNPs with guide RNA specific for the gene coding for blue fluorescent protein (bfp) were immobilized on the platform. As a negative control, RNPs with guide RNA targeting a scramble (nonspecific) sequence were used. The team observed a significantly larger signal with bfp-specific amplicons than with the nonspecific functionalized RNPs. This result indicates that CRISPR-Chip’s signal output is specific for genes complementary to the immobilized RNP complex.
Confident about their platform after the preliminary test, the researchers performed a similar experiment with genomic DNA extracted from HEK cells. They used two types of DNA: one containing the bfp sequence and a control group lacking the sequence. Once again, CRISPR-Chip showed significantly enhanced signal on exposure to the target genomic DNA, relative to control samples lacking the target sequence.
This was a big deal, because they realized their system could detect DNA without amplification. Plus, their assay time was just 15 minutes.
Demonstration of CRISPR-Chip Success in Clinical Samples
Dr. Aran’s ultimate goal of using this platform in diagnostics warranted a sample measurement showing the ability of this system for the same. Therefore, the team tested samples with two commonly deleted exons (exon 3 and 51) in patients suffering from Duchenne Muscular Dystrophy (DMD). Samples from healthy patients were used as a control.
CRISPR-Chip functionalized with RNPs specific to either exon showed significantly enhanced signal output after exposure to genomic samples containing the target exons, relative to DMD samples carrying exon deletions.
Dr. Aran notes that she used Synthego’s single guide RNAs for their experiments.

Overcoming Challenges and Future Steps
The CRISPR-Chip system with its fast detection time and low limit of detection might sound simple, but it is not devoid of challenges.
“Developing this platform requires a very good understanding of nanotechnology and electronics too,” says Dr. Aran. The team had to not only optimize CRISPR guides and RNPs, but they also needed heavy optimization on the electronics side, according to Dr. Aran. For instance, they needed to make sure that they were not absorbing the salts from buffers in their biological solution.
Dr. Aran plans to exercise similar caution when moving forward with her ambitious projects using this platform. Apart from further optimization for diagnostic applications, there are multiple ways to expand this tool in the future for better understanding CRISPR binding. “We can get a lot more information which is actually very useful for pharma companies using CRISPR for therapeutics because we can actually monitor the binding efficiency of their guides and their complex with target DNA,” says Dr. Aran. “So with our sensor and the immobilized CRISPR atop of it, we can actually do a lot of quality control testing, which is the next thing that we are going to do with the sensors.”
Original PublicationHajian, Reza, et al. "Detection of unamplified target genes via CRISPR–Cas9 immobilized on a graphene field-effect transistor." Nature Biomedical Engineering (2019): 1.
Hajian, Reza, et al. "Detection of unamplified target genes via CRISPR–Cas9 immobilized on a graphene field-effect transistor." Nature Biomedical Engineering (2019): 1.
Extra Learning Resources
- What is dead Cas9 and where is it used? Learn more about Cas nuclease variants in this guide.
- Excited about the potential of CRISPR in diagnostics? Check out our podcast episode with Trevor Martin, CEO of Mammoth Biosciences to learn more about CRISPR diagnostics.
- Dr. Aran specifies using RNP format of CRISPR components. What are the benefits of RNPs over other formats?
- What is dead Cas9 and where is it used? Learn more about Cas nuclease variants in this guide.
- Excited about the potential of CRISPR in diagnostics? Check out our podcast episode with Trevor Martin, CEO of Mammoth Biosciences to learn more about CRISPR diagnostics.
- Dr. Aran specifies using RNP format of CRISPR components. What are the benefits of RNPs over other formats?