Mouse Embryo Microinjection Protocol

Contents
In 2019, Dr. David Liu’s lab at Harvard University made a splash in the genome engineering world with prime editing, a brand new gene-editing tool. Prime editing allows for a versatile range of genetic modifications while skipping double-strand breaks and the need for a donor template. It uses a catalytically inactive Cas9 nickase, fused to reverse transcriptase, and a prime editing guide RNA.
Fast forward to 2021, Dr. Joseph M. Miano’s lab at the Augusta University in Georgia published results from their study using prime editing in mice. This is the first study comparing traditional CRISPR-mediated homology directed repair (HDR) with prime editing in vivo.
“Dark Matter of The Genome” and Why it Matters
Dr. Miano has spent the last two decades working on what he refers to as “the dark matter of the genome” or the non-coding DNA. He finds it fascinating to interrogate this regulatory genome using various gene editing tools, including CRISPR editing.
Non-coding DNA (part of DNA that does not code for proteins) accounts for roughly 98% of the total human genome. Conventionally, this was thought of as junk DNA, but scientists are now beginning to understand its role in regulating gene transcription. Transcription, which implies transcribing or copying genetic information from the DNA into a messenger RNA (mRNA), involves proteins called transcription factors (TFs) that bind to the DNA. Transcription factors bind to a stretch of DNA called the transcription factor binding site (TFBS).
Dr. Miano’s lab is interested in a particular kind of TFBSs called CArG boxes. CArG boxes are binding sites for a certain type of transcription factor called serum response factor (SRF).
Using Prime Editing To Study Genetic Changes in Regulatory DNA in Mice
Single nucleotide variants (SNVs) are genetic mutations that have been correlated with several diseases and are common in non-coding DNA, which is also where the TFBS is found. Studying these variants thus provides a deeper understanding of the transcription regulation mechanisms.
Conventional reporter assays and gene regulation experimentsConventionally, the genetic variants in TFBSs have been studied either in cell lines or using Cre/lox reporters in mice. Scientists have also attempted to study SNVs in the native genomic background. However, these experiments involving embryonic stem cell editing cost enormous time and labor. Additionally, these experiments often yield non-specific results, given the genetic redundancy associated with TFBSs, meaning in some cases, TFBSs can be seemingly non-functional. Traditional HDR-based editing is a plausible approach to study SNVs in transcription regulation. The caveats are off-target edits and non-specific insertion-deletions (indels) on the target gene.
Conventionally, the genetic variants in TFBSs have been studied either in cell lines or using Cre/lox reporters in mice. Scientists have also attempted to study SNVs in the native genomic background. However, these experiments involving embryonic stem cell editing cost enormous time and labor. Additionally, these experiments often yield non-specific results, given the genetic redundancy associated with TFBSs, meaning in some cases, TFBSs can be seemingly non-functional. Traditional HDR-based editing is a plausible approach to study SNVs in transcription regulation. The caveats are off-target edits and non-specific insertion-deletions (indels) on the target gene.
Prime editing vs. HDR: the first comparative study in micePrime editing, a more recent technique, does not require a donor template and allows for a wide range of edits, usually with higher precision compared to the traditional HDR editing method. Prime editing can introduce very precise nucleotide substitutions and, unlike base editing, is not limited to transitions (nucleotide base substitutions of similar shapes, either two-ringed purines or one-ringed pyrimidines).
For the first time, Dr. Miano’s lab compared the efficiency and accuracy of prime editing and HDR-based editing in mice. A multi-institutional collaborative endeavor, this study consolidates animal model-based research to understand transcription regulation. It also underpins the impact of single nucleotide variants in non-coding DNA on gene expression.
Prime editing, a more recent technique, does not require a donor template and allows for a wide range of edits, usually with higher precision compared to the traditional HDR editing method. Prime editing can introduce very precise nucleotide substitutions and, unlike base editing, is not limited to transitions (nucleotide base substitutions of similar shapes, either two-ringed purines or one-ringed pyrimidines).
For the first time, Dr. Miano’s lab compared the efficiency and accuracy of prime editing and HDR-based editing in mice. A multi-institutional collaborative endeavor, this study consolidates animal model-based research to understand transcription regulation. It also underpins the impact of single nucleotide variants in non-coding DNA on gene expression.
Genetic Changes in Non-Coding DNA and Gene Expression Regulation
The CArG box sequence is well-conserved across several mammalian species, including mice. In humans, an SRF-binding CArG site is identified upstream of the TSPAN2 gene locus, and the Tspan2 gene codes for a membrane protein in mammals.
Does CArG box regulate Tspan2 gene expression?The authors were curious if the CArG box is essential for Tspan2 gene expression in mice. Interestingly, the transcription start site for Tspan2os, an antisense long non-coding RNA (LncRNA), is located downstream of the Tspan2 start site. So, the authors set out to investigate the role of the CArG box in potentially regulating Tspan2 and Tspan2os transcription in vivo.
Dr. Miano’s team introduced a single base conversion (C>G) to the CArG box using the two-component prime editing approach in mice. Parallel to this, they introduced three-nucleotide substitutions through the conventional HDR editing approach. They injected the CRISPR components into the fertilized mice oocytes.
Both the synthetic guide RNA sequence for HDR editing and the pegRNA sequences were synthesized in collaboration with Synthego.
The authors were curious if the CArG box is essential for Tspan2 gene expression in mice. Interestingly, the transcription start site for Tspan2os, an antisense long non-coding RNA (LncRNA), is located downstream of the Tspan2 start site. So, the authors set out to investigate the role of the CArG box in potentially regulating Tspan2 and Tspan2os transcription in vivo.
Dr. Miano’s team introduced a single base conversion (C>G) to the CArG box using the two-component prime editing approach in mice. Parallel to this, they introduced three-nucleotide substitutions through the conventional HDR editing approach. They injected the CRISPR components into the fertilized mice oocytes.
Both the synthetic guide RNA sequence for HDR editing and the pegRNA sequences were synthesized in collaboration with Synthego.
"I think we had really good fortune with Synthego’s pegRNA, it worked beautifully! I was worried that the prime editor would just continue to polymerize into the scaffold and we would get the edit, but that we would also get some scaffold RNA incorporated into the final DNA template. Well, that didn't happen, and it really worked perfectly, which was astonishing to me and was quite pleasing!"
- Dr. Joseph M. Miano
Single nucleotide change in CArG box impacts Tspan2 expressionThe authors used immuno-RNA fluorescence and in situ hybridization analysis to confirm the loss of Tspan2 mRNA in smooth muscle cells. They observed tissue-specific loss of Tspan2 mRNA expression in mice aorta and bladder with prime editing and HDR experiments. In fact, prime editing of a single nucleotide in the CArG box led to a near-complete loss of expression for both Tspan2 and the overlapping LncRNA, Tspan2os.
The authors used immuno-RNA fluorescence and in situ hybridization analysis to confirm the loss of Tspan2 mRNA in smooth muscle cells. They observed tissue-specific loss of Tspan2 mRNA expression in mice aorta and bladder with prime editing and HDR experiments. In fact, prime editing of a single nucleotide in the CArG box led to a near-complete loss of expression for both Tspan2 and the overlapping LncRNA, Tspan2os.
Prime Editing vs. HDR: Who Takes The Trophy?
The other important question Dr. Miano’s group asked in this study was how prime editing compares to traditional HDR editing in mice. The authors performed targeted sequencing in the founder mice (transgenic mice that have integrated the transgenic construct) to evaluate off-target effects and on-target indels. They used CHANGE-seq analysis to determine off-target effects and targeted sequencing via rhAmp Seq to assess on-target indels.
The results suggested that while the HDR experiments resulted in highly efficient on-target edits in founder mice (55.65%, mean percentage of sequences with correct editing), they also led to unwanted indels on-target (~40%) and off-target edits. On the other hand, the prime editing approach resulted in no spurious indels on the target gene, no off-target effects, and ~21% on-target editing frequency.
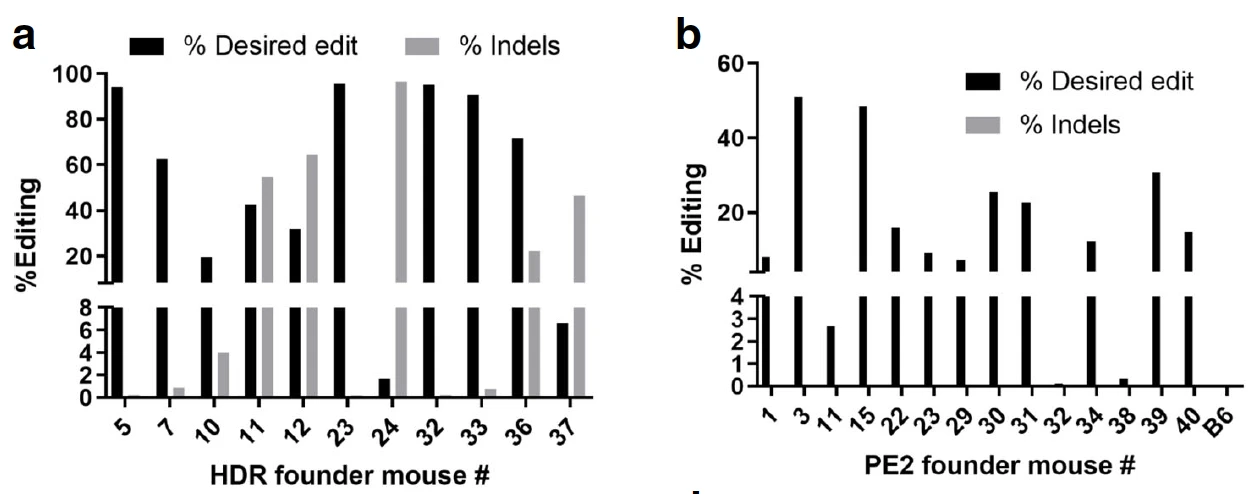
Figure 1. Prime editing shows no on-target indels indicating high precision editing compared to HDR edits. (a) HDR founder mice show high editing efficiency but also show relatively higher spurious indels on the target gene. (b) Prime editing yields efficient editing and no unwanted on-target indels, suggesting higher fidelity than HDR. (Adapted from Fig. 6, Gao et al. Genome Biology (2021) 22:83)
In conclusion, Dr. Miano’s group found that prime editing is a high-precision tool for gene editing in vivo, enabling researchers to study SNVs in non-coding DNA. Importantly, this study enables scientists to develop mice models to understand transcription regulation precisely.
Prime Time For Gene Editing In Vivo
Scientists have long known that single nucleotide variants in non-coding DNA are associated with multiple human diseases. Yet, only a select few of them have been well-studied in animal models of disease progression and fewer even in their native genetic setting. The ability to precisely edit single nucleotide bases using prime editing in vivo enables the creation of robust animal models to elucidate transcription regulation mechanisms. This study will surely pave the way for the future to help us better understand a vital field.