CRISPR is emerging as a widely popular genome editing tool with applications in several areas. Due to its precision and ease, CRISPR gene editing technology has promising potential in therapeutics. However, the CRISPR system sometimes shows off-target effects, i.e., the nuclease cuts at unintended sequences in the genome. The fear of deleterious effects caused by these off-targets limits the widespread adoption of CRISPR in human trials.
Researchers are working on multiple levels to make CRISPR safer. On one hand, they are developing tools to detect CRISPR off-targets, while on the other hand, efforts are ongoing to increase the specificity of the technology itself.
In a step forward in this direction, researchers from Duke University have reported a new approach to make CRISPR editing about 50-fold more precise by adding secondary hairpins to the spacer part of guide RNA.
"CRISPR is specific enough for biological investigations. As far as off-targets go, I think you really want to worry about them when you try and develop a therapeutic,"
says Dr. Dewran Kocak, the first author of this recent study, published in Nature Biotechnology journal.
How Do Hairpin RNAs Reduce CRISPR Off-Targets?
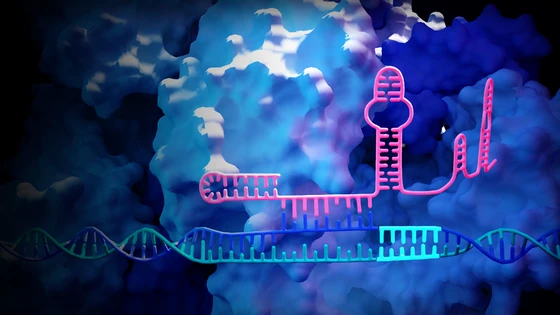
One approach for reducing off-target DNA cleavage is to make it an energetically expensive process. The principle behind introducing an engineered hairpin in the spacer region of a guide RNA is simple. The hairpin is stable in its folded secondary structure conformation. By adjusting the strength of the secondary structure, the researchers could ensure that CRISPR nuclease is active at the desired on-target site but is impeded at undesired off-target sites. When the guide binds to the target site, the DNA-RNA binding is energetically more favorable, so the hairpin unfolds and binding proceeds as usual. However, in the case of off-target binding, the RNA–DNA mispairing is energetically more expensive for hairpin unfolding, and the hairpin will maintain its secondary conformation. This impedes R-loop formation, which is essential for Cas9 activity, at off-target sites.
The authors had the idea of using hairpin-RNAs a few years back, while working on in vitro biochemical characterization of Cas9-DNA binding. While testing the impact of different sgRNA formats, including hairpin structures in the spacer region on nuclease binding, the researchers noted that hairpin (hp) sgRNAs reduced non-target Cas9 binding.
These initial in vitro experiments laid the foundation for the present study. “We started working with the cells to see if it could be extended into human cells to improve specificity,” says Kocak.
Characterization of the Hairpin-sgRNAs with Dead Cas9 In Vivo
Kocak and his team designed RNA hairpins using commercially available software. They needed to verify that any effect on nuclease activity in cells was because of the hairpin structures and not just an effect of the additional RNA sequence. So the researchers tested hp-sgRNAs of varying lengths and non-structured (ns) sgRNAs of similar lengths as control.
They chose the following system for their experiments: dead Cas9-coupled activators targeting the endogenous promoter of IL1RN gene in human cells. Activated gene levels, measured by quantitative PCR, indicated CRISPR activity. The authors observed that increasing hairpin lengths showed decreasing gene activation—a testimony to successful hairpin folding, as this effect was not observed with ns-sgRNAs. Increasing lengths also correlated generally with low free energy for RNA folding.
Hairpin RNAs Increase Specificity of Six Cas Nucleases In Vivo
Next, the researchers tested their hypothesis in a standard CRISPR experiment: they chose the most commonly used nuclease, SpCas9, and used hp-sgRNAs and normal sgRNAs targeting genes with well-characterized off-target sites.
Based on the experiments with dCas9, the team chose hp-sgRNAs of lengths that did not hamper on-target activation, yet showed low enough free energy to fold stably into secondary structures, for their SpCas9 experiments. On indel analysis, the researchers observed fewer off-targets with hp-sgRNAs than with normal guides. The on-target activity was comparable for guides with and without hairpins.
What if adding hp-sgRNAs caused new off-targets? The researchers addressed this question using CIRCLE-Seq, a tool that detects genome-wide off-targets in vitro. They noted that hp-sgRNA showed 124 fewer off-target sites than with the normal guide RNA, and did not generate additional off-targets.
Kocak was not surprised, given the sequential mechanism of Cas9, wherein it first finds the PAM sequence and then unwinds the DNA from 3’ to 5’ end.
"The 3’ end of the guide is the most important thing that determines where Cas9 is going to interact with DNA. If you add things on the 5’e end, you would expect that it would not be as important for directing where Cas9 binds, "
he says.
The authors also performed similar experiments with Cas9 and Cas12 nuclease variants. They found that hp-sgRNAs showed about 55-fold higher specificity compared with normal sgRNAs across 6 different Cas variants. This study lays the groundwork for testing engineered hairpins to reduce off-targets in clinically relevant samples.
“One thing we'd really like to see is this used in some kind of therapeutically relevant disease model. Our lab specializes in specifically investigating therapies for DMD, Duchenne Muscular Dystrophy. I'm working with a couple of my other lab members on that front to develop some more specific guide RNAs for those kinds of therapies,” says Kocak about their future plans.
Original PublicationKocak, D. Dewran, et al. "Increasing the specificity of CRISPR systems with engineered RNA secondary structures." Nature Biotechnology (2019): 1.
Kocak, D. Dewran, et al. "Increasing the specificity of CRISPR systems with engineered RNA secondary structures." Nature Biotechnology (2019): 1.
Image CreditsHeader Image: Ella Maru
Header Image: Ella Maru