What is an Experimental Control?
In the scientific world, we are always excited to see what type of results are produced from our experiments. These results can have a range of impacts, from serving as key results in a manuscript to groundbreaking discoveries that save lives. However, these results are only effective if appropriate controls are included in the experiments for comparison. So what is an experimental control?
Experimental controls are standards with predictable outcomes that are tested alongside actual samples under the same conditions to validate the experimental design and set up. For example, while testing for gene expression using PCR, primers targeting a known constitutively expressed gene are run as a positive control while a negative control can be just the primers without the DNA sample. Running controls helps researchers verify that the success or failure of their experiment is real and not caused by unknown complications, as those would result in irregular and unexpected control readouts.
The introduction of CRISPR has resulted in the expansion of genome editing research for studying medicine, agriculture, biofuels, and diagnostics. With such a drastic increase in data and growth, using controls to gauge if and how well the genome editing experiment worked is just as essential as the actual outcome of the experiment.
When performing CRISPR-based genome editing experiments in cells, it is essential to know what types of controls should be used to understand if the Cas nuclease actually made it into the cells AND if a CRISPR edit actually occurred. For example, if you want to create a CRISPR knockout cell line, what types of controls would you need to consider? Or if you are observing no editing with your sgRNA of interest, what controls would help determine if the issue is inefficient delivery or inefficient guide design?
Types of CRISPR Controls
For a CRISPR gene knockout experiment to be successful, first and foremost, all of the CRISPR components, namely the guide RNA and Cas nuclease, need to be delivered inside of the cells. However, unoptimized workflow conditions may impact the delivery of CRISPR components into the cells resulting in inefficient or no genome editing. So how do you test if your transfection worked?
Transfection controlsThe majority of the time, the CRISPR components are colorless making it difficult to see if they made it inside of the cells. Being able to visualize and quantify if the material is entering your cells can help determine if the CRISPR components have been delivered successfully. Using a transfection control is one way you can quantify and visually check if the delivery is successful.
Transfection controls can consist of a fluorescence reporter such as green fluorescent protein mRNA or plasmid to be the reference point if the material is being delivered inside of cells. Please note, you can use other fluorescent proteins or markers that can be successfully observed if they are in or outside of the cell. Using transfection protocols such as lipofection, electroporation, or nucleofection that can be found on the Synthego Website, you can set up a transfection workflow starting with using a transfection control to assess how effectively a fluorescence reporter was delivered inside of a cell. A low fluorescence detected in the cells following transfection implies that the reporter may not be entering the cells. This would require optimizing your workflow conditions such as changing the concentration of the reporter being used, adjust the amount of lipofection reagents you are using, check for expired reagents, adjust parameters on your electroporators, changing the cell density, etc.
If you would like further suggestions on how to troubleshoot your workflow conditions, you can always reach out to Synthego’s Tech Support for additional tips and recommendations.
While transfection controls are essential to assess transfection efficiency and identify if the material is entering the cells and to what extent, how do you know the CRISPR components will cut DNA?
The majority of the time, the CRISPR components are colorless making it difficult to see if they made it inside of the cells. Being able to visualize and quantify if the material is entering your cells can help determine if the CRISPR components have been delivered successfully. Using a transfection control is one way you can quantify and visually check if the delivery is successful.
Transfection controls can consist of a fluorescence reporter such as green fluorescent protein mRNA or plasmid to be the reference point if the material is being delivered inside of cells. Please note, you can use other fluorescent proteins or markers that can be successfully observed if they are in or outside of the cell. Using transfection protocols such as lipofection, electroporation, or nucleofection that can be found on the Synthego Website, you can set up a transfection workflow starting with using a transfection control to assess how effectively a fluorescence reporter was delivered inside of a cell. A low fluorescence detected in the cells following transfection implies that the reporter may not be entering the cells. This would require optimizing your workflow conditions such as changing the concentration of the reporter being used, adjust the amount of lipofection reagents you are using, check for expired reagents, adjust parameters on your electroporators, changing the cell density, etc.
If you would like further suggestions on how to troubleshoot your workflow conditions, you can always reach out to Synthego’s Tech Support for additional tips and recommendations.
While transfection controls are essential to assess transfection efficiency and identify if the material is entering the cells and to what extent, how do you know the CRISPR components will cut DNA?
Controls for CRISPR editing experimentsUsing a validated control of a standard target region that cuts DNA using CRISPR can help optimize workflow conditions to ensure the highest editing efficiency is achieved either before conducting or in parallel to the main CRISPR experiments. Genotyping after editing using a CRISPR experimental control can help assess how well the transfection conditions are optimized to perform a CRISPR edit.
Experimental controls involve using the CRISPR components in various ways to assess the editing of the DNA. Positive editing controls, negative editing controls, and mock controls are just a few examples of CRISPR controls that can be used in different ways to help assess the editing efficiency and how well workflow conditions are optimized.
Using a validated control of a standard target region that cuts DNA using CRISPR can help optimize workflow conditions to ensure the highest editing efficiency is achieved either before conducting or in parallel to the main CRISPR experiments. Genotyping after editing using a CRISPR experimental control can help assess how well the transfection conditions are optimized to perform a CRISPR edit.
Experimental controls involve using the CRISPR components in various ways to assess the editing of the DNA. Positive editing controls, negative editing controls, and mock controls are just a few examples of CRISPR controls that can be used in different ways to help assess the editing efficiency and how well workflow conditions are optimized.
Positive editing controlA positive editing control consists of a validated guide RNA that has been demonstrated to have high editing efficiencies. In many human cell models, a common target region of a genome is genes such as TRAC, RELA, and CDC42BPB. One of the common mouse genes used as a target region for positive controls is ROSA26. All of these controls can be purchased from Synthego’s CRISPRevolution Add-Ons page. In most cases, an editing positive control has been proven to be effective at making DNA cuts when workflow conditions are optimized across a wide variety of cell lines and transfections conditions. This control is essential to verify the transfection conditions are optimized based on the high editing efficiency observed. One effective way to assist with determining the editing efficiency is by using Synthego’s Interference of CRISPR Edit, or ICE for short, as it is a free online bioinformatics tool that can assist with assessing the editing efficiency of Sanger sequenced CRISPR samples.
A positive editing control consists of a validated guide RNA that has been demonstrated to have high editing efficiencies. In many human cell models, a common target region of a genome is genes such as TRAC, RELA, and CDC42BPB. One of the common mouse genes used as a target region for positive controls is ROSA26. All of these controls can be purchased from Synthego’s CRISPRevolution Add-Ons page. In most cases, an editing positive control has been proven to be effective at making DNA cuts when workflow conditions are optimized across a wide variety of cell lines and transfections conditions. This control is essential to verify the transfection conditions are optimized based on the high editing efficiency observed. One effective way to assist with determining the editing efficiency is by using Synthego’s Interference of CRISPR Edit, or ICE for short, as it is a free online bioinformatics tool that can assist with assessing the editing efficiency of Sanger sequenced CRISPR samples.
Negative editing control
A negative editing control should not create a genomic edit to establish a baseline of how the cell reacts to the stress induced by transfection workflows. In most cases, the phenotypes observed using the editing negative control should be comparable to that of the wildtype cell line. Further details of a few types of editing negative controls that can be used are given below.
1. Scramble guide RNA and Cas NucleaseScramble guide RNA are guide RNA that do not have a complementary sequence in the genome to direct a Cas nuclease to cut the genome.
Scramble guide RNA are guide RNA that do not have a complementary sequence in the genome to direct a Cas nuclease to cut the genome.
2. Guide RNA OnlyRemoving the Cas nuclease and only delivering guide means the component that cuts DNA is not present. Therefore, no CRISPR edits will occur. The guide RNA used can be the same guide RNA used for the CRISPR editing experiment.
Removing the Cas nuclease and only delivering guide means the component that cuts DNA is not present. Therefore, no CRISPR edits will occur. The guide RNA used can be the same guide RNA used for the CRISPR editing experiment.
3. Cas Nuclease OnlyRemoving the guide RNA and only delivering Cas nuclease means the Cas nuclease cannot be direct to any location in the genome to make a CRISPR edit.
Synthego also offers a few negative editing controls that can be found on the CRISPRevolution Add-Ons page. Utilizing an editing negative control is essential because it provides insight if the phenotype you are observing when performing your CRISPR edit experiments is a true phenotype or if the phenotype observed is caused by the cellular response of the cells when put under the stress of transfection.
Removing the guide RNA and only delivering Cas nuclease means the Cas nuclease cannot be direct to any location in the genome to make a CRISPR edit.
Synthego also offers a few negative editing controls that can be found on the CRISPRevolution Add-Ons page. Utilizing an editing negative control is essential because it provides insight if the phenotype you are observing when performing your CRISPR edit experiments is a true phenotype or if the phenotype observed is caused by the cellular response of the cells when put under the stress of transfection.
Mock controlThe last type of control that could be used is a mock control, wherein cells are transfected without Cas nuclease and guide RNA. The cells are still going through the stressful transfection conditions induced by electroporation or lipofection but nothing is being delivered inside of the cells. In most cases, the phenotype observed from the mock control should be similar to the wildtype. Therefore in a similar way to the editing negative controls, a mock control will provide insight into if the phenotype observed from the CRISPR edit experiment is a true phenotype or from the cellular response the cell creates when exposed to transfection conditions.
With so many different types of controls, it can be overwhelming to understand when to use them. However, they are essential to have to set up CRISPR experiments.
The last type of control that could be used is a mock control, wherein cells are transfected without Cas nuclease and guide RNA. The cells are still going through the stressful transfection conditions induced by electroporation or lipofection but nothing is being delivered inside of the cells. In most cases, the phenotype observed from the mock control should be similar to the wildtype. Therefore in a similar way to the editing negative controls, a mock control will provide insight into if the phenotype observed from the CRISPR edit experiment is a true phenotype or from the cellular response the cell creates when exposed to transfection conditions.
With so many different types of controls, it can be overwhelming to understand when to use them. However, they are essential to have to set up CRISPR experiments.
What Steps Should Include CRISPR Controls?
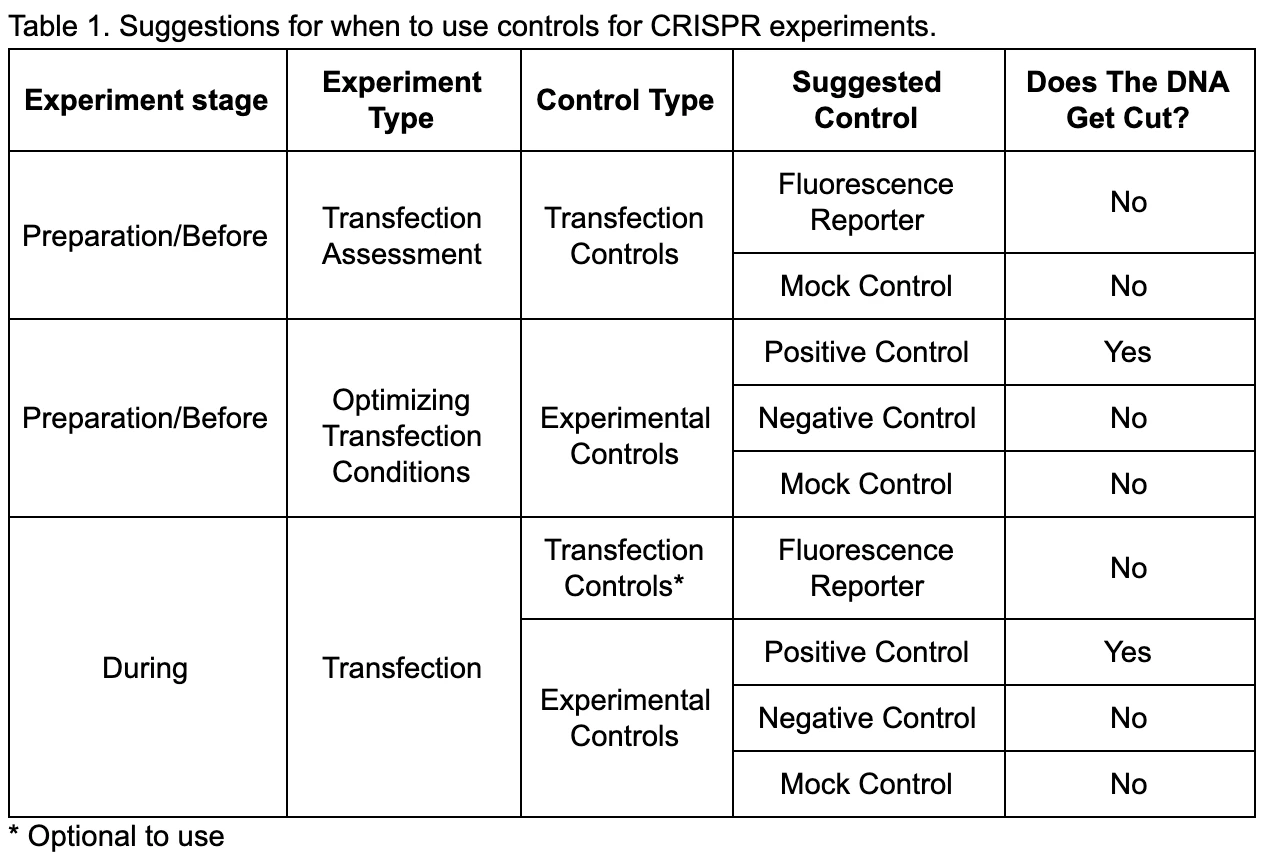
Using appropriate controls is essential to understand how well your CRISPR edit experiments worked. The control can be used for optimizing transfection conditions to ensure material is entering the cells and if the highest editing efficiency is reached when these optimized conditions are tweaked perfectly. It is also important to use controls in parallel when you are proceeding through the main CRISPR experiments to use as a comparison to confirm genome editing is occurring correctly. The following table contains suggestions of various steps when you use these controls in your CRISPR genome editing workflow.
Secure Controls for CRISPR Experiments
At Synthego, we offer a wide variety of controls for your CRISPR experiments through our CRISPRevolution Add-Ons order page. Our controls have been tested and validated for common transfection methods in a variety of mouse and human cell lines to ensure they will be effective for a wide range of CRISPR-based workflows. There is even a multi-guide control for the human Gene Knockout v2 Kits to assist with optimizing transfection conditions.
The genome-editing world is utilizing CRISPR across broad fields to provide data to help society understand the world around us and help improve our living. Unless you have a control to compare your results to or controls used to optimize workflow conditions, the data becomes difficult to interpret or validate impacting how we effectively can communicate results. Selecting the correct controls and using these controls to ensure all conditions are optimized can provide a foundation for successful CRISPR experiments that will make an impact on the world.