With the recent approval of Casgevy (exa-cel), the first CRISPR-based medicine, and a swathe of new CRISPR therapies progressing through clinical trials, the effects of CRISPR off-target editing are now very much in the spotlight. In this blog, we’re covering how CRISPR off-target edits occur and why they’re a problem, how to predict and analyze the off-target effects of CRISPR, strategies to minimize off-targets, and how Synthego’s approach can help you develop the safest CRISPR therapies.
Understanding CRISPR Off-Target Editing
First, let’s look at the basics of CRISPR off-targets: what are they, how do they occur, and why are they such a problem?
What is off-target CRISPR editing?CRISPR off-target editing refers to the non-specific activity of the Cas nuclease at sites other than the intended target, causing undesirable or unexpected effects on the genome. The majority of CRISPR off-target edits occur at known sites that bear homology to the target sequence. In some cases, even on-target CRISPR editing can have unwanted consequences, such as chromosomal translocations or chromothripsis.
CRISPR off-target editing refers to the non-specific activity of the Cas nuclease at sites other than the intended target, causing undesirable or unexpected effects on the genome. The majority of CRISPR off-target edits occur at known sites that bear homology to the target sequence. In some cases, even on-target CRISPR editing can have unwanted consequences, such as chromosomal translocations or chromothripsis.
How does off-target CRISPR editing occur?Wild-type CRISPR systems have a reasonable level of tolerance for mismatches between their target sequence and their guide RNA (gRNA), making them… a little promiscuous. In fact, the wild-type Cas9 from Streptococcus pyogenes (SpCas9) can tolerate between three and five base pair mismatches, meaning that it can potentially create double-stranded breaks at a range of sites in the genome if they bear similarity to the intended target and have the correct PAM sequence.
Wild-type CRISPR systems have a reasonable level of tolerance for mismatches between their target sequence and their guide RNA (gRNA), making them… a little promiscuous. In fact, the wild-type Cas9 from Streptococcus pyogenes (SpCas9) can tolerate between three and five base pair mismatches, meaning that it can potentially create double-stranded breaks at a range of sites in the genome if they bear similarity to the intended target and have the correct PAM sequence.
Why is CRISPR off-target editing a problem?
The off-target effects of CRISPR are a concern because they can confound the results of your experiments and decrease repeatability. However, the level of risk depends largely on the intended application of your CRISPR experiments and where the off-target edits occur. If an off-target edit occurs in a non-coding region of the genome, like an intron, it may not cause any problems. In contrast, if it occurs within the protein-coding regions of a gene, it can have a considerable impact.
Let’s take a functional genomics application as an example. If you’re trying to determine the function of a specific gene in a cell line or organism via CRISPR knockout, off-target CRISPR activity can make it difficult to determine if the observed phenotype is the result of the intended edit or the off-target activity.
If your aim is to create a cell or gene therapy for use in humans, CRISPR off-target editing becomes a much more significant concern. In a clinical setting, the off-target effects of CRISPR negatively impact the results of clinical trials and cause delays in the clinical development pipeline. More importantly, they also pose critical safety risks to patients; if an off-target edit causes a mutation to arise in an oncogene, there could be life-threatening consequences.
Even within the clinical applications of CRISPR, there are varying levels of risk associated with off-target editing. For cell therapies, where CRISPR editing is performed ex vivo, individual cells can be selected, lowering the risk. For gene therapy applications, in which editing occurs in vivo, extra care must be taken to ensure no off-target effects will occur because they cannot be selected for or reversed once the treatment is delivered to the patient.
During the review process of Casgevy - a CRISPR cell therapy designed to treat sickle cell disease - the FDA focused on the potential for the medicine to cause off-target edits, highlighting that people who carry rare genetic variants may be at higher risk. The FDA guidance on human genome editing now states that preclinical and clinical studies should include characterization of CRISPR off-target editing to minimize potential safety concerns.
The off-target effects of CRISPR are a concern because they can confound the results of your experiments and decrease repeatability. However, the level of risk depends largely on the intended application of your CRISPR experiments and where the off-target edits occur. If an off-target edit occurs in a non-coding region of the genome, like an intron, it may not cause any problems. In contrast, if it occurs within the protein-coding regions of a gene, it can have a considerable impact.
Let’s take a functional genomics application as an example. If you’re trying to determine the function of a specific gene in a cell line or organism via CRISPR knockout, off-target CRISPR activity can make it difficult to determine if the observed phenotype is the result of the intended edit or the off-target activity.
If your aim is to create a cell or gene therapy for use in humans, CRISPR off-target editing becomes a much more significant concern. In a clinical setting, the off-target effects of CRISPR negatively impact the results of clinical trials and cause delays in the clinical development pipeline. More importantly, they also pose critical safety risks to patients; if an off-target edit causes a mutation to arise in an oncogene, there could be life-threatening consequences.
Even within the clinical applications of CRISPR, there are varying levels of risk associated with off-target editing. For cell therapies, where CRISPR editing is performed ex vivo, individual cells can be selected, lowering the risk. For gene therapy applications, in which editing occurs in vivo, extra care must be taken to ensure no off-target effects will occur because they cannot be selected for or reversed once the treatment is delivered to the patient.
During the review process of Casgevy - a CRISPR cell therapy designed to treat sickle cell disease - the FDA focused on the potential for the medicine to cause off-target edits, highlighting that people who carry rare genetic variants may be at higher risk. The FDA guidance on human genome editing now states that preclinical and clinical studies should include characterization of CRISPR off-target editing to minimize potential safety concerns.
Regulatory Approval of CRISPR Therapies eBook
CRISPR cell and gene therapies allow clinicians to address the root cause of genetic disease. However, taking these complex biologics from discovery to delivery requires both scientific and regulatory expertise. This eBook explores the CRISPR regulatory space, from clinical trials to regulatory frameworks, and Synthego’s regulatory support for your CRISPR-based therapeutic journey.

Predicting, Detecting, and Analyzing CRISPR Off-Target Editing
Even outside of translational research, peer reviewers will often expect you to test for off-target events. If off-target editing does occur, you may need to demonstrate that it has not impacted the results. Let’s explore methods for CRISPR off-target prediction, detection, and analysis.
CRISPR off-target predictionWhen selecting your gRNAs for a CRISPR experiment, there are often multiple guides to choose from, and it’s crucial to select a guide with low similarity to other sites in the genome. Guide design software, such as CRISPOR, will help you choose a guide using specialized algorithms for CRISPR off-target prediction.
For each gRNA, design tools will typically offer a CRISPR off-target score or ranking based on the predicted on-target to off-target activity; high-ranking gRNAs will have high on-target activity and a lower risk of off-target editing. CRISPR off-target prediction is crucial because the candidate sites for off-target editing will inform your approach to detection and analysis after you perform editing experiments.
When selecting your gRNAs for a CRISPR experiment, there are often multiple guides to choose from, and it’s crucial to select a guide with low similarity to other sites in the genome. Guide design software, such as CRISPOR, will help you choose a guide using specialized algorithms for CRISPR off-target prediction.
For each gRNA, design tools will typically offer a CRISPR off-target score or ranking based on the predicted on-target to off-target activity; high-ranking gRNAs will have high on-target activity and a lower risk of off-target editing. CRISPR off-target prediction is crucial because the candidate sites for off-target editing will inform your approach to detection and analysis after you perform editing experiments.
CRISPR off-target detection and analysisThere are several strategies for the detection of off-target CRISPR editing. One of the most common is to simply sequence the predicted off-target sites, identified during gRNA selection. This is often referred to as candidate site sequencing and can be representative of the total number of off-target edits – if there are very few off-target editing events at these candidate sites, it’s relatively unlikely there will be off-target edits at sites with low similarity.
Another approach is to use targeted sequencing. These methods sequence specific sites in the genome, such as any site that has been bound by the Cas protein, or any site where NHEJ has occurred. Some examples include Cas CHIP-seq, GUIDE-seq, CIRCLE-seq, DISCOVER-seq, and TEG-seq. Another approach, known as CAST-seq, was designed specifically to identify and quantify chromosomal rearrangements as a result of CRISPR editing.
Whole genome sequencing is the only way to perform a full and comprehensive analysis of CRISPR off-target editing, including the detection of chromosomal aberrations. However, it is significantly more expensive, making it less common than targeted sequencing methods.
When it comes to analyzing CRISPR experiments, the Inference of CRISPR Edits (ICE) tool is ideal for discovery-stage research. Cited in more than 400 publications, ICE offers free, fast, and robust analysis of overall editing efficiencies as well as CRISPR off-target edits. Using Sanger sequencing data and the chosen guide sequence, you can assess your editing efficiencies simultaneously in large batches and select cells for clonal isolation. Producing publication-quality figures, ICE is compatible with any species and CRISPR edit.
There are several strategies for the detection of off-target CRISPR editing. One of the most common is to simply sequence the predicted off-target sites, identified during gRNA selection. This is often referred to as candidate site sequencing and can be representative of the total number of off-target edits – if there are very few off-target editing events at these candidate sites, it’s relatively unlikely there will be off-target edits at sites with low similarity.
Another approach is to use targeted sequencing. These methods sequence specific sites in the genome, such as any site that has been bound by the Cas protein, or any site where NHEJ has occurred. Some examples include Cas CHIP-seq, GUIDE-seq, CIRCLE-seq, DISCOVER-seq, and TEG-seq. Another approach, known as CAST-seq, was designed specifically to identify and quantify chromosomal rearrangements as a result of CRISPR editing.
Whole genome sequencing is the only way to perform a full and comprehensive analysis of CRISPR off-target editing, including the detection of chromosomal aberrations. However, it is significantly more expensive, making it less common than targeted sequencing methods.
When it comes to analyzing CRISPR experiments, the Inference of CRISPR Edits (ICE) tool is ideal for discovery-stage research. Cited in more than 400 publications, ICE offers free, fast, and robust analysis of overall editing efficiencies as well as CRISPR off-target edits. Using Sanger sequencing data and the chosen guide sequence, you can assess your editing efficiencies simultaneously in large batches and select cells for clonal isolation. Producing publication-quality figures, ICE is compatible with any species and CRISPR edit.
Strategies to Minimize CRISPR Off-Target Editing
Anyone who has worked with CRISPR systems knows that creating their intended edit and avoiding off-target effects requires the perfect balance between the Cas nuclease, the gRNA, and the delivery vehicle. Here, we’ll take a look at how these different strategies affect the likelihood of off-target editing.
Choosing the right Cas nuclease
As we mentioned earlier, the most commonly used SpCas9 nuclease has a reasonable risk of off-target editing. Fortunately, there is now a wide range of different Cas nucleases available to choose from depending on your application. Many of these alternatives, such as Cas12 or Cas13, have different off-target activity from SpCas9.
Spin-offs and derivatives of CRISPR technology, such as base editing, prime editing, and epigenome editing, can also reduce the likelihood of off-target editing because they do not create DSBs in the genome. These technologies can use catalytically dead Cas9 (dCas9) to simply bind DNA and direct the activity of other proteins, or a Cas9 nickase (nCas9) to create single-stranded breaks. In the case of nCas9, a dual-guide approach can produce a similar effect to creating a DSB while reducing the chance of off-target editing.
If creating DSBs is necessary, there are several versions of Cas9, including high-fidelity variants, that have been designed or engineered to have lower off-target activity. Unfortunately, despite their lower off-target activity, many high-fidelity Cas9 nucleases come with the drawback of having reduced on-target editing, making them less than ideal for certain applications.
It’s also important to note that high-fidelity nucleases have reduced off-target cleavage, but not necessarily reduced off-target DNA binding. For example, if you’re using a high-fidelity catalytically dead Cas9 nuclease (dCas9) to perform epigenetic editing, the likelihood of off-target epigenetic editing will not be reduced because this involves binding of the Cas enzyme to the DNA, rather than target cleavage.
As we mentioned earlier, the most commonly used SpCas9 nuclease has a reasonable risk of off-target editing. Fortunately, there is now a wide range of different Cas nucleases available to choose from depending on your application. Many of these alternatives, such as Cas12 or Cas13, have different off-target activity from SpCas9.
Spin-offs and derivatives of CRISPR technology, such as base editing, prime editing, and epigenome editing, can also reduce the likelihood of off-target editing because they do not create DSBs in the genome. These technologies can use catalytically dead Cas9 (dCas9) to simply bind DNA and direct the activity of other proteins, or a Cas9 nickase (nCas9) to create single-stranded breaks. In the case of nCas9, a dual-guide approach can produce a similar effect to creating a DSB while reducing the chance of off-target editing.
If creating DSBs is necessary, there are several versions of Cas9, including high-fidelity variants, that have been designed or engineered to have lower off-target activity. Unfortunately, despite their lower off-target activity, many high-fidelity Cas9 nucleases come with the drawback of having reduced on-target editing, making them less than ideal for certain applications.
It’s also important to note that high-fidelity nucleases have reduced off-target cleavage, but not necessarily reduced off-target DNA binding. For example, if you’re using a high-fidelity catalytically dead Cas9 nuclease (dCas9) to perform epigenetic editing, the likelihood of off-target epigenetic editing will not be reduced because this involves binding of the Cas enzyme to the DNA, rather than target cleavage.
Alternatives to CRISPR-Cas9: Nucleases for Next-Gen Therapy
There are now many alternatives to CRISPR-Cas9. In this blog, we explore CRISPR alternative nucleases and how they are being used to create new therapies.
Optimizing gRNA design
The simplest strategy to minimize CRISPR off-target editing is to design your gRNAs carefully. As we discussed earlier, design tools will rank all possible gRNAs for your target site based on their on-target/off-target ratio. In proof-of-concept experiments, it's common to select several of the top-ranking gRNAs and test all of them in cell lines, since the top-ranked guide in silico may not produce the best results in a biological model.
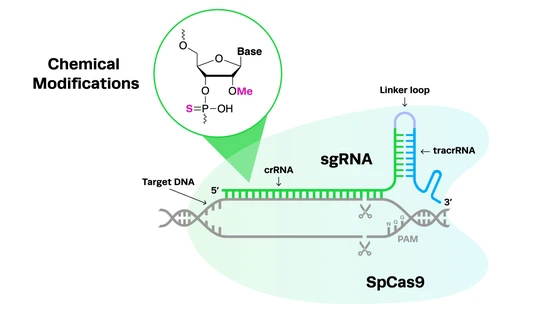
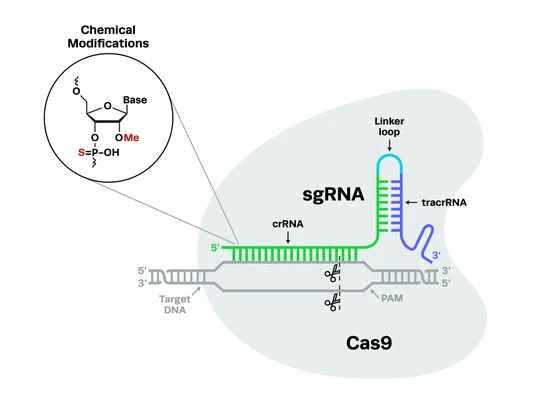
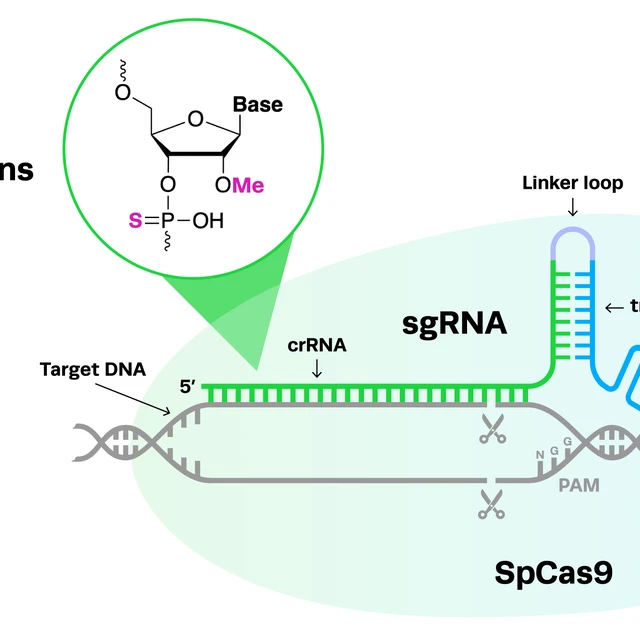
For any CRISPR experiments, particularly in vivo applications, it's important to consider the addition of chemical modifications to your gRNAs. Chemical modifications on gRNAs have many benefits including reducing the likelihood of CRISPR off-target editing. At Synthego, we often add 2'-O-methyl analogs (2'-O-Me) and 3' phosphorothioate bond (PS) modifications to our synthetic gRNAs to reduce off-target edits and increase editing efficiency at the target site. For more detail on chemical modifications, check out our detailed blog on this topic below.
GC content is another key consideration when designing your gRNAs. Higher GC content in the gRNA sequence stabilizes the DNA:RNA duplex when the guide binds to the target, increasing on-target editing and reducing off-target binding. Guide length can also influence the likelihood of off-target CRISPR editing: shorter gRNAs of 20 nucleotides or less have a lower risk of off-target activity.
The simplest strategy to minimize CRISPR off-target editing is to design your gRNAs carefully. As we discussed earlier, design tools will rank all possible gRNAs for your target site based on their on-target/off-target ratio. In proof-of-concept experiments, it's common to select several of the top-ranking gRNAs and test all of them in cell lines, since the top-ranked guide in silico may not produce the best results in a biological model.
For any CRISPR experiments, particularly in vivo applications, it's important to consider the addition of chemical modifications to your gRNAs. Chemical modifications on gRNAs have many benefits including reducing the likelihood of CRISPR off-target editing. At Synthego, we often add 2'-O-methyl analogs (2'-O-Me) and 3' phosphorothioate bond (PS) modifications to our synthetic gRNAs to reduce off-target edits and increase editing efficiency at the target site. For more detail on chemical modifications, check out our detailed blog on this topic below.
GC content is another key consideration when designing your gRNAs. Higher GC content in the gRNA sequence stabilizes the DNA:RNA duplex when the guide binds to the target, increasing on-target editing and reducing off-target binding. Guide length can also influence the likelihood of off-target CRISPR editing: shorter gRNAs of 20 nucleotides or less have a lower risk of off-target activity.
Demystifying CRISPR gRNA Chemical Modifications
Confused about chemical modifications on CRISPR guides? Join us as we explore why chemical modifications are so important, the different types and their applications, and how Synthego’s modified guides can support your clinical success.
Selecting the best CRISPR cargo and delivery vehicleThe type of CRISPR cargo and delivery vehicle you choose also have a significant impact on the likelihood of off-target editing because they affect how long the CRISPR components will be active within cells. The longer CRISPR components stay active, the more chance they have of creating off-target cuts. This makes the short-term expression of gene editing components highly desirable to reduce CRISPR off-target editing in clinical applications.
The type of CRISPR cargo and delivery vehicle you choose also have a significant impact on the likelihood of off-target editing because they affect how long the CRISPR components will be active within cells. The longer CRISPR components stay active, the more chance they have of creating off-target cuts. This makes the short-term expression of gene editing components highly desirable to reduce CRISPR off-target editing in clinical applications.
CRISPR cargoIf your CRISPR components – the Cas nuclease and guide – take the form of a DNA plasmid cargo, it will take some time for the nuclease and guide sequences to be transcribed into mRNA and then for the nuclease to be translated into protein, delaying the start of editing. This cargo format also causes the components to stick around in the cells for a while, and therefore carries an increased risk of causing off-target effects.
If the cargo takes the form of a ribonucleoprotein (RNP) complex, in which the guide is an RNA molecule and the Cas nuclease is a protein, there is no need for transcription or translation, speeding up the initial editing process. In this format, the components will typically display high levels of expression immediately after delivery, followed by rapid decay and clearance. This gives them much less time to cause off-targets and has the added advantage of reducing other potential safety risks, such as activating immune responses.
If your CRISPR components – the Cas nuclease and guide – take the form of a DNA plasmid cargo, it will take some time for the nuclease and guide sequences to be transcribed into mRNA and then for the nuclease to be translated into protein, delaying the start of editing. This cargo format also causes the components to stick around in the cells for a while, and therefore carries an increased risk of causing off-target effects.
If the cargo takes the form of a ribonucleoprotein (RNP) complex, in which the guide is an RNA molecule and the Cas nuclease is a protein, there is no need for transcription or translation, speeding up the initial editing process. In this format, the components will typically display high levels of expression immediately after delivery, followed by rapid decay and clearance. This gives them much less time to cause off-targets and has the added advantage of reducing other potential safety risks, such as activating immune responses.
CRISPR delivery vehiclesThe delivery vehicle you package these cargoes in can also determine the likelihood of CRISPR off-target editing. If your cargo is delivered in an integrating lentiviral vector, it will be integrated into the host genome and expressed long-term, significantly increasing the chances of off-target editing. In contrast, if the cargo is packaged in a lipid nanoparticle (LNP), there is no risk of integration and the components will degrade rapidy.
The delivery vehicle you package these cargoes in can also determine the likelihood of CRISPR off-target editing. If your cargo is delivered in an integrating lentiviral vector, it will be integrated into the host genome and expressed long-term, significantly increasing the chances of off-target editing. In contrast, if the cargo is packaged in a lipid nanoparticle (LNP), there is no risk of integration and the components will degrade rapidy.
CRISPR Delivery Methods: Cargo, Vehicles, and Challenges
Efficient CRISPR delivery is essential in cell and gene therapies. In this article, we discuss the many different CRISPR delivery methods, including viral, non-viral, and physical.
Synthego’s Commitment to Creating Safer Genomic Medicines
At Synthego, we are committed to providing best-in-class gene editing reagents to our customers and making CRISPR medicines as safe as possible. With these goals in mind, we have developed several partnerships and products that allow our clients to reduce the risk of CRISPR off-target editing in their therapeutic candidates.
SeQure Dx: Off-target editing analysis for clinical applicationsIn order to provide high-quality off-target analysis for our clinical customers, Synthego has partnered with leading off-target analysis company SeQure Dx. As we mentioned earlier, designing and choosing gRNAs is a crucial first step in minimizing the likelihood of CRISPR off-targets. Our partnership with SeQure Dx simplifies the process of choosing the right guides, characterizing your final gRNA molecule, and filing for Investigational New Drug (IND) status with the FDA.
SeQure Dx’s proprietary methods for off-target analysis are nuclease-agnostic, allowing for the use of any CRISPR system, and can be customized for your experiments. Importantly, this mode of analysis takes into account sequence variants from more than 4000 human genomes and can incorporate your own variant data if required. They are also designed to meet the latest guidelines from regulators like the FDA, streamlining the preparation of regulatory documents.
In order to provide high-quality off-target analysis for our clinical customers, Synthego has partnered with leading off-target analysis company SeQure Dx. As we mentioned earlier, designing and choosing gRNAs is a crucial first step in minimizing the likelihood of CRISPR off-targets. Our partnership with SeQure Dx simplifies the process of choosing the right guides, characterizing your final gRNA molecule, and filing for Investigational New Drug (IND) status with the FDA.
SeQure Dx’s proprietary methods for off-target analysis are nuclease-agnostic, allowing for the use of any CRISPR system, and can be customized for your experiments. Importantly, this mode of analysis takes into account sequence variants from more than 4000 human genomes and can incorporate your own variant data if required. They are also designed to meet the latest guidelines from regulators like the FDA, streamlining the preparation of regulatory documents.
hfCas12Max: A powerful high-fidelity nuclease for CRISPR therapeuticsIn addition to traditional SpCas9, Synthego now offers a next-generation nuclease designed for clinical applications: high-fidelity Cas12Max (hfCas12Max). Unlike high-fidelity Cas9 variants, this small but powerful nuclease exhibits low off-target editing while retaining high on-target activity. With broad PAM recognition but high specificity, hfCas12Max produces staggered-end or ‘sticky’ DSBs, making it an attractive nuclease for knock-in approaches. Adding to its many advantages, hfCas12Max acts with shorter guide molecules, further reducing the possibility of producing off-target edits. Ideal for in vivo or ex vivo CRISPR medicines, we offer commercial sub-licensing for this nuclease to our partners.
In addition to traditional SpCas9, Synthego now offers a next-generation nuclease designed for clinical applications: high-fidelity Cas12Max (hfCas12Max). Unlike high-fidelity Cas9 variants, this small but powerful nuclease exhibits low off-target editing while retaining high on-target activity. With broad PAM recognition but high specificity, hfCas12Max produces staggered-end or ‘sticky’ DSBs, making it an attractive nuclease for knock-in approaches. Adding to its many advantages, hfCas12Max acts with shorter guide molecules, further reducing the possibility of producing off-target edits. Ideal for in vivo or ex vivo CRISPR medicines, we offer commercial sub-licensing for this nuclease to our partners.
eSpot-ON: An engineered high-fidelity Cas9 with a superior safety profileFurther expanding the CRISPR toolkit, Synthego is now commercializing eSpot-ON, an engineered version of a naturally occurring Cas9 variant, which comes as either recombinant protein or mRNA format. Despite having naturally high on-target editing and low off-target activity, eSpot-ON was further engineered to be as safe as possible for the development of CRISPR therapeutics. eSpot-ON is so named because of its high-fidelity gene editing activity; it has higher on-target editing and lower off-target editing than any other HiFi SpCas9 currently available. Producing staggered-end DSBs in DNA, it can be used for both HDR and NHEJ. This incredible nuclease has an excellent safety profile, reducing the risks associated with gene editing for clinical applications, including chromosomal translocations.
Further expanding the CRISPR toolkit, Synthego is now commercializing eSpot-ON, an engineered version of a naturally occurring Cas9 variant, which comes as either recombinant protein or mRNA format. Despite having naturally high on-target editing and low off-target activity, eSpot-ON was further engineered to be as safe as possible for the development of CRISPR therapeutics. eSpot-ON is so named because of its high-fidelity gene editing activity; it has higher on-target editing and lower off-target editing than any other HiFi SpCas9 currently available. Producing staggered-end DSBs in DNA, it can be used for both HDR and NHEJ. This incredible nuclease has an excellent safety profile, reducing the risks associated with gene editing for clinical applications, including chromosomal translocations.
Unlocking the IP Benefits of Novel Nucleases in CRISPR Therapies
Explore the intellectual property benefits of incorporating novel nucleases into CRISPR therapies in our latest article. Discover how improved licensing models, strategic IP protections, and expanded gene target access can transform the landscape for biotech investors, researchers, and scientists.

The CRISPR field continues to evolve, with significant progress in the detection and reduction of off-target editing. At Synthego, we look forward to a future where CRISPR therapies are equitable and safe for all patients who need them.
Hopefully this blog helped you further your understanding of CRISPR off-target editing and offered some strategies to reduce these unwanted effects in your research. If you enjoy reading this type of technical content, be sure to subscribe to The Bench Blog!