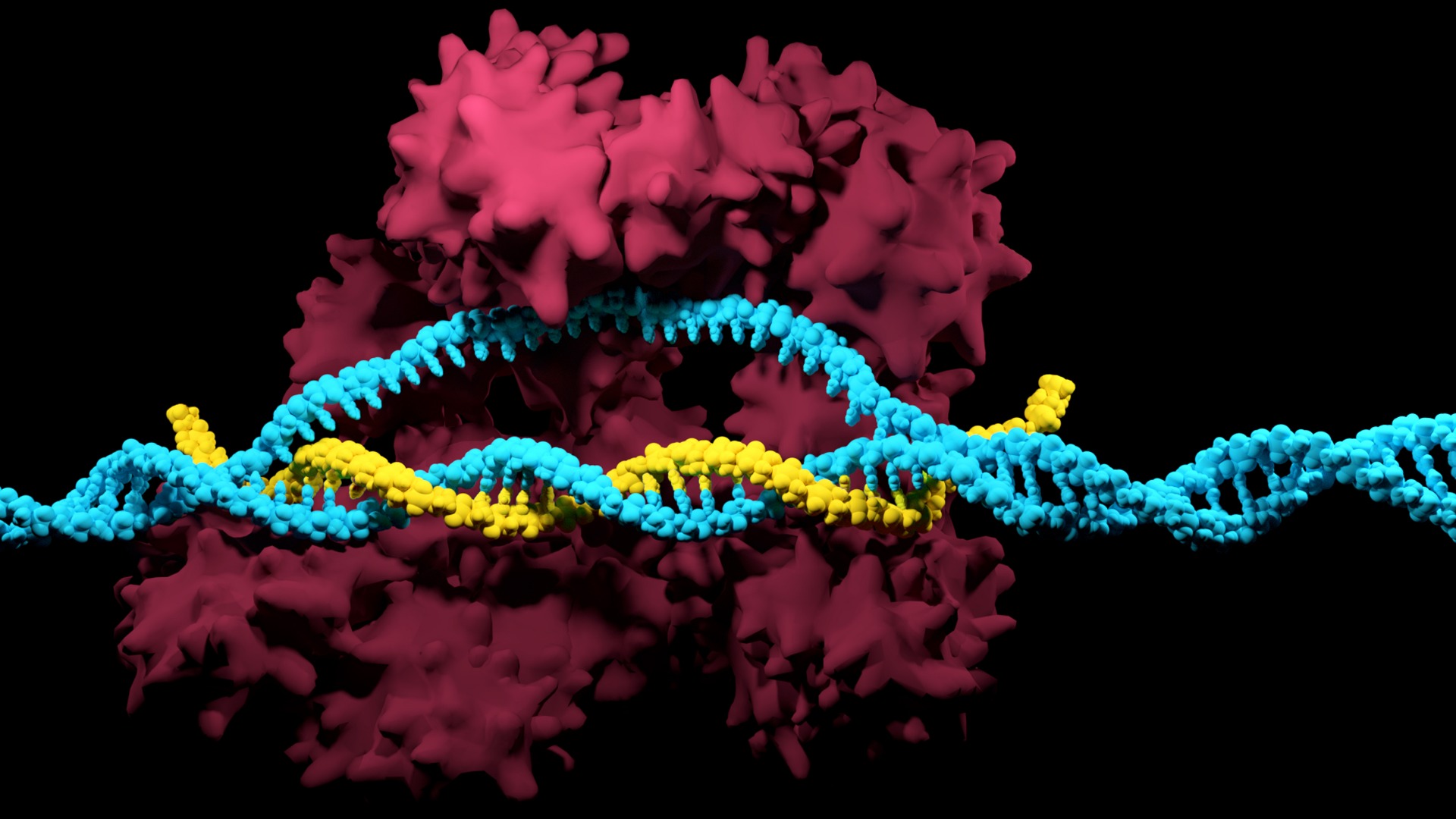
CRISPR-Cas9 is a simple two-component system that allows researchers to precisely edit any sequence in the genome of an organism. This is achieved by guide RNA, which recognizes the target sequence, and the CRISPR-associated endonuclease (Cas) that cuts the targeted sequence.
Researchers across the globe who are adopting this technology are bound to come across an important term: PAM sequence. What is a PAM? How is it involved in CRISPR genome engineering? Why is it important in designing a CRISPR experiment?
In this article, we answer all these questions to clarify all PAM-related doubts and enable you to successfully venture into the world of genome engineering.
What is the Protospacer Adjacent Motif (PAM) Sequence?
The protospacer adjacent motif (or PAM for short) is a short DNA sequence (usually 2-6 base pairs in length) that follows the DNA region targeted for cleavage by the CRISPR system, such as CRISPR-Cas9. The PAM is required for a Cas nuclease to cut and is generally found 3-4 nucleotides downstream from the cut site.
To better understand the Cas dependence on this protospacer adjacent motif, let’s first dive into the mechanism of CRISPR. Today, CRISPR is being used in all types of organisms and has countless applications. But before scientists harnessed the CRISPR system to serve as a genome engineering tool, it served an important purpose in nature.
The whole purpose of the CRISPR system is to protect bacteria from invading viruses (bacteriophage). When a virus attacks bacteria, the cells mount a primitive immune response. Any bacteria that survive infection keep a part of the viral DNA as a way to remember the past infection so they can fight off the virus easier next time. This is achieved by the Cas1 and Cas2 nucleases, which work together as a complex to identify the invading virus. Once Cas1 and Cas2 have done their job, Cas9 then cuts out a segment of the virus, known as a protospacer, which is added to the CRISPR array between the sequence repeats.
In the future, if the same virus invades again, the bacterial cell now has a complementary RNA sequence which recruits the Cas nuclease to cut the viral DNA and stop the infection. But with this system in place, how does the viral genome fragment embedded in the bacterial genome not trigger the Cas9 to cut the bacterial genome?
Enter PAM. The PAM, also known as the protospacer adjacent motif, is a short specific sequence following the target DNA sequence that is essential for cleavage by Cas nuclease.
The PAM is about 2-6 nucleotides downstream of the DNA sequence targeted by the guide RNA and the Cas cuts 3-4 nucleotides upstream of it. In S. pyogenes, for example, Cas9 recognizes a 5′-NGG-3′ PAM (where “N” can be any nucleotide base). However, the spacers in its CRISPR array are coded by 5’-GTT-3’, so the Cas9 cannot cut the bacteria’s own genome.
Another critical function of PAM is that the Cas nuclease will search for it before unravelling the viral DNA in order to cut. When Cas identifies the correct PAM, it will then check to see if the upstream region matches the guide RNA before it makes the edit.
CRISPR 101 eBook
CRISPR has quickly become a standard laboratory tool for gene editing. As the adoption of CRISPR accelerates worldwide, up-to-date knowledge of the basics of CRISPR is essential for anyone in the field. From target identification studies to the recent breakthroughs in clinical trials, CRISPR is enabling scientists to unlock the power of the genome.
Download our CRISPR 101 eBook today to stay up to date on all your CRISPR basics and get the best results in your CRISPR experiments!
Download
To Learn More About CRISPR And Its Various Applications, Explore Our CRISPR 101 eBook
Role of PAM in CRISPR Experiment Design
The genomic locations that can be targeted for editing by CRISPR are limited by the presence and locations of the nuclease-specific PAM sequences. There are several factors/workarounds that scientists can consider to expand their target sites while designing their genome engineering experiments.
Different PAMs for Different Cas Nucleases
The most commonly-used Cas9 from Streptococcus pyogenes, also referred to as SpCas9, recognizes the PAM sequence 5′-NGG-3′ (where “N” can be any nucleotide base). What if the target genomic locus does not bear this NGG sequence? Researchers cannot afford to be limited by the presence of the specific PAM for their CRISPR experiments.
Thankfully, there are many different Cas endonucleases to choose from isolated from different bacterial species, and each recognizes a different PAM. For instance, hfCas12Max, an engineered high-fidelity Cas12 variant, PAM sequence is 5'-TN and/or 5'-TNN and SaCas9 (from Staphylococcus aureus) specifically recognizes the short PAM sequence 5′-NNGRR(N)-3. There have also been studies identifying S. pyogenes Cas9 mutants that have altered specificity or recognize novel PAM sequences. Other Cas nucleases, including Cas9 from other species, recognize longer PAMs or have higher specificity to minimize off-target cutting. So researchers have the option of selecting another nuclease whose corresponding PAM may be present in the target genome. Different nucleases may also offer additional advantages, making the choice of nuclease a crucial step in the CRISPR experiment.
Table 1. Summary of Cas and other nuclease variants used in CRISPR experiments and their PAM sequences.
| CRISPR Nucleases |
Organism Isolated From |
PAM Sequence (5’ to 3’) |
|
SpCas9
|
Streptococcus pyogenes |
NGG |
|
hfCas12Max
|
Engineered from Cas12i |
TN and/or TNN |
| SaCas9 |
Staphylococcus aureus |
NNGRRT or NNGRRN |
| NmeCas9 |
Neisseria meningitidis |
NNNNGATT |
| CjCas9 |
Campylobacter jejuni |
NNNNRYAC |
| StCas9 |
Streptococcus thermophilus |
NNAGAAW |
| LbCpf1 (Cas12a) |
Lachnospiraceae bacterium |
TTTV |
| AsCpf1 (Cas12a) |
Acidaminococcus sp. |
TTTV |
| AacCas12b |
Alicyclobacillus acidiphilus |
TTN |
| BhCas12b v4 |
Bacillus hisashii |
ATTN, TTTN and GTTN |
| Cas14 |
Uncultivated archea |
T-rich PAM sequences, eg. TTTA for dsDNA cleavage, no PAM sequence requirement for ssDNA |
| Cas3 |
in silico analysis of various prokaryotic genomes |
No PAM sequence requirement |
The ingenuity of researchers has also enabled the CRISPR community to not rely on naturally available enzymes but even engineer them as per their need. SpCas9, for example, has been engineered to recognise PAM sequences other than NGG. There are several papers using directed evolution to achieve altered PAM specificity.
Role of PAM in Guide RNA Design
The very basis of bacteria evading their own endonucleases is the PAM sequence. When the Cas nuclease cuts fragments of the viral genome and stores them in the CRISPR repeats, it excludes the PAM sequence even though it is involved in recognition to ensure that the bacterial genome is not recognized as a target.
Thus, following this logic, when researchers design a gRNA sequence for their CRISPR experiments, they generally exclude the PAM from the guide RNA. This is especially important for plasmid-based systems, as the delivery format is DNA the region encoding the gRNA will be chopped by the Cas if it also contains the PAM sequence.
However, there are new applications of CRISPR surfacing which requires researchers to contradict this logic and include the PAM sequence in their design.
Homing Guide RNAs Include the PAM
A new method of cellular barcoding was introduced recently that allows researchers to track cellular differentiation. This mechanism involved designing guide RNAs in which the PAM sequence is intentionally included so they could target themselves for cleavage.
The concept of homing RNAs was introduced by part of the same team in a previous publication a couple of years ago. Consider the basic CRISPR mechanism in bacteria: the DNA sequences that encode guide RNAs are not cleaved by the Cas nuclease themselves because they do not contain PAM sequences. The researchers reverse-engineered this concept and developed homing RNAs, which target their own DNA sequence for cleavage, simply by adding in the PAM sequence in the guides. Their previous study has shown that homing guide RNA (hgRNA) editing results in far more diversity in accumulated mutations relative to conventional CRISPR guides. This barcoding technique allows researchers to trace the lineage of cells through development. Read our blog post for further applications of homing RNAs.
While the guide RNAs and the Cas nucleases steal the limelight for all discussions about CRISPR, the PAM sequence is equally important. If the target DNA regions to be engineered do not have a PAM, then editing simply will not occur.
Understanding PAMs and how they work will give you a leg up on the competition and ensure that you’re able to take full advantage of CRISPR technology in your research.
A Multiplexed High Throughput Method to Evaluate Novel Nuclease Activity
Synthego's expertise and Paragon Genomics technology have been combined to develop a new method to evaluate novel nucleases that enables direct comparison of their editing activity. This tech note takes a closer look at this collaboration displaying the power of Synthego's Halo™ Platform for Synthetic sgRNA and automation capabilities coupled with Paragon Genomics CleanPlex™ amplicon sequencing technology to have a side-by-side comparison of the activity of nucleases. Data is provided.
Download Tech Note
Take A Deep Dive Into A Method Developed To Evaluate Editing Efficiency Between Different Nucleases
Understanding PAM Sequences Ensures Successful Editing
We hope this chapter helped you understand the importance of the PAM sequence and the PAM recognition requirements of the various Cas nucleases so that you can take full advantage of CRISPR technology in your research. While the guide RNAs and the Cas nucleases steal the limelight for all discussions about CRISPR, the PAM sequence is equally important. If the target DNA regions to be engineered do not have a PAM, then editing simply will not occur.
The discovery of more novel Cas nucleases with different PAM recognitions and stringencies will open new doors for CRISPR gene editing technology. As you will read in our next chapter, How to Choose the Right Cas Variant for Every CRISPR Experiment, many Cas nucleases have been, or are currently being, engineered to achieve altered PAM specificity. These developments will enable a wider range of targets for gene editing and also improve accuracy and reduce off-target effects of CRISPR, which are crucial if it is to be used for gene therapy in humans.