Contents
CRISPR is well known for its powerful potential in therapeutics but this tool does more than gene editing. CRISPR’s search function is being utilized in diagnostics—the latest example being the newly-invented CRISPR-Chip for target DNA detection.
Today, we chat with Omar Abudayyeh and Jonathan Gootenberg, co-founders of Sherlock Biosciences, one of the top CRISPR start-up companies already making news in the medical diagnostic industry. This dynamic duo is famous for discovering the Cas13 enzyme, developing the SHERLOCK CRISPR diagnostic kit, and balancing academia and their own company.
Learn more about their journey from grad student life to co-founding Sherlock Biosciences in this interview.
Listen to the podcast here, or jump down and start learning & reading the podcast transcript — the choice is yours!
How CRISPR Diagnostics is Changing Disease Detection
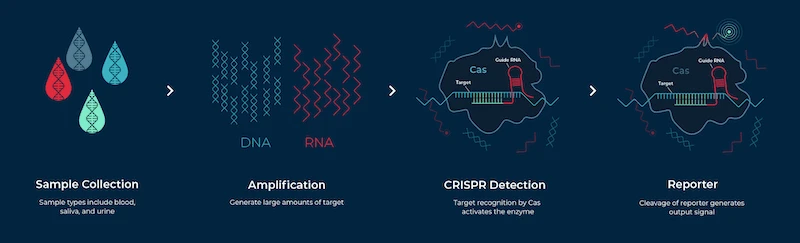
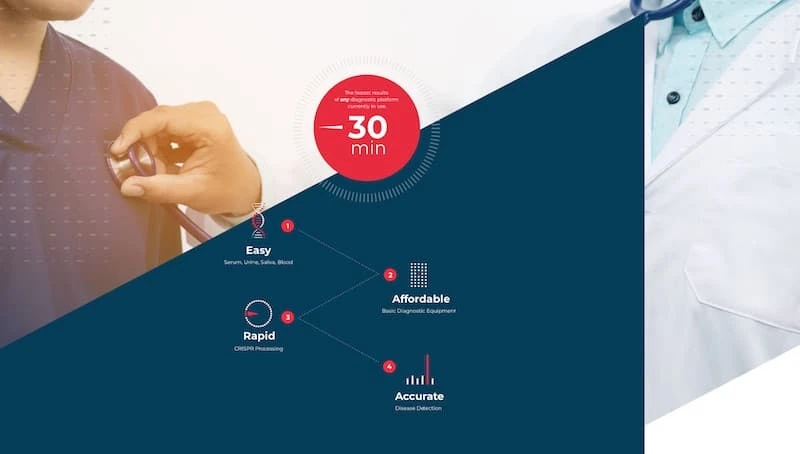
Traditional microbiological diagnostic methods such as PCR are time consuming assays that require multiple steps and dedicated equipment. In contrast, CRISPR-based assays require minimal to no equipment and can be run as single reaction tests. CRISPR diagnostic tests are also easy to use, and can deliver fast and accurate results.
What is a CRISPR diagnostic test?A CRISPR diagnostic test harnesses the “search” function of CRISPR nucleases to detect DNA or RNA mutations associated with a disease. Several researchers are leveraging this unique property of CRISPR to develop diagnostic kits that can detect diseases using patient samples such as blood, urine, or saliva.
A CRISPR diagnostic test harnesses the “search” function of CRISPR nucleases to detect DNA or RNA mutations associated with a disease. Several researchers are leveraging this unique property of CRISPR to develop diagnostic kits that can detect diseases using patient samples such as blood, urine, or saliva.
How do CRISPR diagnostic tests work?A CRISPR system typically consists of two components––an endonuclease and a guide RNA (gRNA). The gRNA locates the specified DNA sequence and the Cas nuclease then makes a double stranded cut at the target location. For diagnostic applications, the most popularly used nucleases are Cas12 and Cas13.
A CRISPR system typically consists of two components––an endonuclease and a guide RNA (gRNA). The gRNA locates the specified DNA sequence and the Cas nuclease then makes a double stranded cut at the target location. For diagnostic applications, the most popularly used nucleases are Cas12 and Cas13.
4 Benefits of CRISPR disease detection testsCRISPR diagnostic tests have many advantages over the traditional assays, including:
- Single reaction - Traditional assays such as PCR require multiple steps, while CRISPR-based diagnostic assays can be run as a single reaction test.
- Minimal equipment - CRISPR diagnostics require minimal to no equipment.
- Speed - These tests can be run quickly.
- Accuracy - CRISPR diagnostic tests are more accurate than traditional tests.
- Easy to use - Because of the minimal equipment necessary, CRISPR diagnostics could be made available as handheld devices.
CRISPR diagnostic tests have many advantages over the traditional assays, including:
- Single reaction - Traditional assays such as PCR require multiple steps, while CRISPR-based diagnostic assays can be run as a single reaction test.
- Minimal equipment - CRISPR diagnostics require minimal to no equipment.
- Speed - These tests can be run quickly.
- Accuracy - CRISPR diagnostic tests are more accurate than traditional tests.
- Easy to use - Because of the minimal equipment necessary, CRISPR diagnostics could be made available as handheld devices.
3 Key Takeaways from this Podcast About SHERLOCK
In the podcast, Abudayyeh and Gootenberg talk about the Cas13 nuclease and how it’s used in SHERLOCK. Here are some of the main points you don’t want to miss out on:
1. The discovery of Cas13 nucleaseThe discovery of Cas13 nuclease was the breakthrough that lead to the development of SHERLOCK.
Time in podcast: 2:34
The discovery of Cas13 nuclease was the breakthrough that lead to the development of SHERLOCK.
Time in podcast: 2:34
2. Cas13 can be used to detect both DNA and RNACas13 can be used to detect both DNA and RNA. During the amplification stage, a T7 promoter can be added to convert DNA into RNA, which is then detected by Cas13.
Time in podcast: 7:24
Cas13 can be used to detect both DNA and RNA. During the amplification stage, a T7 promoter can be added to convert DNA into RNA, which is then detected by Cas13.
Time in podcast: 7:24
3. CRISPR diagnostic kits will soon be available to the general publicWhile an exact timeline is hard to predict, Abudayyeh and Gootenberg hope to have the CRISPR diagnostics kits available soon. They say that the technology is easy to design and test, which makes it easier for things to progress rapidly.
Time in podcast: 11:13
While an exact timeline is hard to predict, Abudayyeh and Gootenberg hope to have the CRISPR diagnostics kits available soon. They say that the technology is easy to design and test, which makes it easier for things to progress rapidly.
Time in podcast: 11:13
Abudayyeh & Gootenberg’s Background & Educational Interests
Learn about Abudayyeh and Gootenberg’s educational backgrounds, how they met, and their research projects.
Omar Abudayyeh: I did my undergrad at MIT a while ago in mechanical engineering and biological engineering. I went in very interested in working with biomedical devices and biotechnology, and that led me to doing an M.D./Ph.D. program at Harvard. I did the first year as a medical school student, and then completed my Ph.D. about a year ago from MIT. I have now put my medical training on hold to start a lab with Jonathan at MIT to continue tackling questions surrounding gene therapy and gene editing technology development.
Jonathan Gootenberg: I also did my undergrad at MIT — that's actually how Omar and I met. I was studying biological engineering and math. Then I went to Harvard to get my Ph.D. in systems biology, and I graduated about eight months ago. Since then, we've been running this new lab where we are exploring new questions in biological diversity.
Minu: So, both of you met in undergrad, and that's when you realized you liked working on similar things. Was it planned to work together later or did that just happen?
Omar & Jonathan: We didn't plan to both join Feng Zhang’s lab, but we had worked together on a senior design project that was actually a fun, crazy idea about engineering bacteria-like probiotics for some inflammatory disease, and it was a fun opportunity to work together. We actually continued doing some research together after that on new microscopy modalities. I think it was pretty fortuitous that we both ended up in Feng's lab.
Minu: You are now also working at the McGovern Research Institute, where both of you have your own labs. Could you tell us a little bit about other projects that you are working on or are interested in?
Omar & Jonathan: I think both of us are pretty interested in furthering gene editing and gene delivery technology. Gene editing still has limitations in that it's not efficient in all cell types, especially brain cells. Current gene editing tools still can't insert DNA well either. I think a lot of our work is looking at technologies that go beyond CRISPR, that can manipulate even more efficiently in more tissues, so that more diseases can be targeted.
Beyond that, we're also interested in a second problem with gene therapy which is that even if you have the perfect genome editor, it would still be hard to deliver these tools to the right tissue and right cells, and have them go in efficiently. We're currently trying to explore certain new types of viruses and other types of modalities that can better get nucleic acid into cells for that purpose. Those are our main two technology development approaches.
Beyond that, we're also understanding how using these tools to probe aging biology, and understand why cells become dysfunctional, and why they become senescent. If we can understand that better, we can determine if we can use our tools to actually reverse senescence and regenerate tissues across multiple aging-associated diseases.
Minu: And for a fun question, because you have such an interesting name of your company and your kit, between the two of you, who is Sherlock, and who is Watson?
Jonathan: I think the better question is who is Moriarty? The important thing about having a good working relationship is that we both have to have flexibility with our roles. It depends on the day, of course, but in general, it's probably half-half.
Minu: So it depends on the day?
Omar: I guess I have more of the medical background, so maybe I'm closer to Watson in that regard. I think the more important thing is what's the next acronym that we can think up because that was certainly the hardest part of SHERLOCK — figuring that out! There were a lot of different iterations it went through. I think it's really important to have fun with the day-to-day; that's what makes it a great experience.
Minu: Absolutely. I think you guys have done a great job, and you've certainly set the bar for abbreviations really high! Every CRISPR researcher now has to step up to that level if they want to come up with any abbreviations. We are looking forward to what your next one will be!
CRISPR Diagnostics Gets Unlocked with SHERLOCK
How was Cas13 discovered? What is it and how can it be used to develop a diagnostic platform?
Minu: One of the main things in your project was the discovery of Cas13, which was fundamental to the development of SHERLOCK. Can you talk a little bit about how you came to find this enzyme and how that led you to where you are now?
Omar & Jonathan: SHERLOCK is our highly-sensitive and specific CRISPR diagnostics platform. It is really exciting that CRISPR can be used for sensing nucleic acids with implications for disease detection, but we definitely didn't go into graduate school interested in diagnostics. I was much more excited about molecular tool development, gene therapy, and treating diseases.
As a part of that, both of us were really interested in exploring the diversity of CRISPR enzymes, so everyone was really excited about Cas9 six years ago. We had this question about what other enzymes in the CRISPR world existed, and if there are any that might have unique features that could be useful for things we hadn’t even conceived of yet.
We did a lot of bioinformatics to find the CRISPR system we characterized biochemically in bacteria. One of the really interesting ones was this Cas13 enzyme. It was unique because it targeted RNA instead of DNA, and that was fairly unique because the enzymes known at the time that we used for those tools were all DNA-targeted. Being able to go after RNA allows you to do so much: you can modulate genes, and you can edit residues on genes temporally so it's like a temporary gene modification.
We later realized you could use this for diagnostics as well, because when this enzyme is bound to its target, it would not only cleave its target, but it would also cleave other molecules in the solution. That property is kind of like saying "hey, I just found my target and I'm just going to cleave everything and tell everyone that I found this target." It's sort of a more abstract way of thinking about it.
We realized that we could put in these fluorescent reporters. When they would cleave they would become fluorescent, and this Cas13 would basically release fluorescence when they found a nucleic acid target. We were also able to make this test sensitive to a single molecule, so it was very quick and cheap. We went down this path very fortuitously from just asking these questions about CRISPR biology, and not really knowing what we would find. And we took that path to many different areas — including CRISPR diagnostics.
Minu: I remember reading about SHERLOCK for the first time, and then I think about a year later, there was an upgraded version which was even more sensitive. What struck me most was that the readout is so simple. Is it correct that the method is so sensitive that it can actually use strips in your sample, and you can actually see a readout on the strip?
Omar & Jonathan: Yes, that's correct. One of the great things about this CRISPR diagnostic platform is that by changing the piece of RNA or DNA that is cleaved by this activity that Omar talked about, we can change it from a readout by fluorescence to something that could be read out by a paper strip. There are also a few who have done it so it actually changes the optical properties of a fluid or even potentially electronic readouts. So, this kind of lateral flow paper-based diagnostic is really powerful because the modularity of the platform allows you to swap the readouts depending on the application.
It was just really fun to think about different ways to develop things. That's the cool thing about being in the lab and having these enzymes. It's really thrilling first to discover them. I remember when we first characterized Cas12, back when it was called Cpf1. It's like people thought there was only Cas9 and now, this is an entirely new thing.
That’s the discovery aspect, but even beyond that, once we have the technologies, you can kind of let your imagination run wild, and ask “what can we detect?” Can we detect cancer? Can we detect different patient mutations, viral DNA, bacterial DNA? Or, can we detect using a different color metric or other readouts? And the thing you don't see, of course, is there are so many things that we tried that failed. But you just have to keep trying and letting your imagination play with it. It can make really interesting stuff like this lateral flow readout.
Minu: That's a very interesting point, especially about the part of failing. In the end, you always see these amazing Science/Nature papers, and it's hard to imagine the hard work that has gone into it, and all the trial and error behind it. You're absolutely right. Just keeping at it is probably the best way to go about it.
Can you speak a little bit about the implications of detecting RNA? One thing is that a lot of viruses have RNA genomes. Where else could it be used to detect RNA instead of DNA?
Omar & Jonathan: Just to clarify, we can actually do both RNA and DNA detection. One of the important steps of this detection reaction is we do a pre-amplification before the Cas13 detection. If you are just doing Cas13 detection, it would get in the low, maybe picomolar-high femtomolar range of sensitivity, which is millions of molecules per microliter. But if we combined in the reaction a set of enzymes that can actually amplify the nucleic acids in solution a little bit, it can bring up a single molecule or ten molecules to a level where Cas13 can then detect it.
What we essentially do is we can turn RNA or DNA into a lot of DNA during amplification. Then in amplification, we add a T7 promoter so that we can take the amplified DNA and make it into RNA, and then the RNA is detected by Cas13. All of that works in one reaction, and it's very quick and very simple at one temperature. What it allows us to do is detect DNA and RNA.
To answer your question where RNA detection is also useful, there are a lot of RNA viruses, bacteria have RNA signatures, 16S-rRNA sequences, or even transcripts of antibiotic resistance genes. Cancers are also really interesting at the RNA level. A lot of transcripts are fused; genes become fused together and you can detect those in the RNA. Hopefully, in general, RNA signatures are better than DNA signatures because they are going to be at a higher copy number, so it's actually easier to detect them per cell or with whatever you're doing.
Sherlock Biosciences: Using CRISPR to Create Diagnostic Testing Solutions
How is owning a start-up different from working in academia? Learn about Abudayyeh and Gootenberg’s perspectives here, and the amazing work their company is doing for the world of diagnostics.
Minu: Now that you have this great technology, you have started a new company: SHERLOCK Biosciences. Can you speak a little about what your roles will be in this company, and how you balance working in a lab and having a company of your own? It's a very interesting position to be in!
Omar & Jonathan: For us, it's a lot of fun to do the early academic development of these technologies when you publish really fantastic papers and get to talk about them at conferences. But, I think there's always that missing piece where you want to be able to say “what's next?” How do we take this work and really make an impact on society for people and patients?
It's hard to be in academia because of the capital that's needed to really scale and translate technology into a product. Because of that, I think getting to spin this off as a company has been a really rewarding experience; we get to interact with different types of people who are thinking about the business of how you actually get the capital to make a product that can make the company sustainable, and we work with people who think about how you make a test that's reliable and accurate across hundreds of patients.
It's one thing to have a proof of concept in the lab, but when you're deploying something that can actually inform how doctors make decisions about people's care and affect patient lives, you have to be sure that these tests are completely reliable, and will give you the right answer—every time.
It's definitely not easy work, but we've assembled a really good team. There are nine co-founders, including Feng Zhang and Jim Collins who originally helped us develop the technology when we were in their labs. We've brought on some other really awesome experts like David Walt who is a co-founder of Illumina, and has really been helping craft the business vision aspect of the diagnostics company. We have a fantastic CEO, Rahul Dhanda, who has 20 years of diagnostics experience. We have a really good team around this that, hopefully, can bring this to patients soon.
Minu: I hope so too, and best of luck with that!
When Will CRISPR Diagnostic Kits be Available to the General Public?
CRISPR diagnostic kits will be brought to market soon. Compared to CRISPR therapeutics, diagnostics face lesser technical, ethical, and regulatory hurdles. While an exact timeline might be hard to predict, it’s safe to say it won’t be long before CRISPR diagnostics kits are available to the general public.

Minu: If you had to estimate, when do you think these diagnostic kits would be available in the market, and, would they be available for just anyone to buy, or will you be sending them to doctors only?
Omar & Jonathan: The future vision will be easy accessibility, because one of the really powerful aspects of the technology is that we can do testing wherever. Of course, there are many different iterations of the technology, and each of those has to go through different aspects of regulatory approval. I think that we will probably initially focus on not giving them directly to patients.
As for the timeline, that's always hard to predict, but we're moving quickly. We’re hoping to have products quite soon; because the technology is so easy to design and test, it’s easier to get something out rapidly.
Another thing, of course, is that the technology is not only being adopted by the company, but there's many different applications across human health, agriculture, and even the environment. We have collaborators who are interested in using it to track different ecological populations, for example, fish migrating on the West Coast or looking at different types of beetles.
It's really exciting to see how a foundational technology like this RNA detection can really be adopted by so many people to make a difference in so many areas.
Where is CRISPR Diagnostic Tech Going in the Next 5 Years?
CRISPR technology is rapidly progressing––new enzymes are being discovered and new techniques are being developed. What is the perception of the general public? Are people enthusiastic and optimistic about this new technology?
Minu: Recently, in the last one or two years especially, there have been so many CRISPR-related companies, and CRISPR research is accelerating. One of the things I was curious about was that CRISPR itself is fairly new - it's less than a decade old - so how do you think the next five years are going to change this field?
Omar & Jonathan: Of course, as this technology becomes more and more available, the applications will be more and more impressive. One great thing about having all of these different methods out there is that it really creates a versatile toolbox for people to use in biology. That will be quite amazing to see.
I think it's always great to bet on biodiversity being more and more surprising. I think we'll continue to find new enzymes and new techniques, and those will probably allow us to do things that we haven't done before, such as more efficient integration into non-dividing cells and other aspects.
Lastly, I think that as people become more familiar with the techniques, the efficiency will certainly go up. As these new techniques are coming through, they always involve an aspect of debugging. If you look at the original Cas9 genome engineering paper, the editing efficiencies are actually quite low compared to what you might expect today.
Of course back then, they were incredibly transformative, but you can just see how a technology - once it has its initial debut - matures, becoming better and better over time. We'll see that maturation of these technologies, and people getting higher and higher efficiencies with editing across more complex models as well. It's really an exciting time, and I think that there is much, much more to be discovered.
Minu: It is absolutely an exciting time! You mentioned people getting more used to the technology. In the scientific community, sure, they'll make even more developments. On the public side, I don't know if you have had the chance to talk about your technology with the general public or non-scientists, but, if you have, can you tell us a little bit about how people are perceiving this technology and if you think that will improve with time or, are they already excited about this?
Omar & Jonathan: We've given some talks to general audiences before, and I think most people leave feeling inspired and excited about the technology. It's probably because they are sort of interested from the perspective of treating disease. I think CRISPR has so much potential to treat disease and actually cure people of diseases that are really nasty. It also goes beyond healthcare where agriculture can make farming much easier with better crop yields.
I think in general when people hear those applications, they feel pretty good about it. I think more recently there have been a lot of questions with things like the CRISPR babies in China and what's going on with that. “Why are people upset about it and what does it mean?” I think we find ourselves explaining that type of situation and what the difference is between gene editing and treating diseases.
I also think one component of it is that a lot of people don’t appreciate the difference between the fact that if you treat someone's disease, you're not actually propagating genetic information that will go to someone's kids. Part of solving this is just explaining what all these CRISPR companies are actually doing, and how it can make a really big difference. It's not going to do anything that's ethically dubious.
Overall, in our experience, people seem pretty optimistic. But, there is definitely explaining we have to do.
Resources to Learn More About CRISPR Diagnostics
The use of CRISPR technology for diagnostic applications is proving to be an exciting field. Several start-up companies are developing CRISPR based diagnostic tools. If you want to learn more about CRISPR diagnostics, you can check out some of the resources below.
Broad Institute video explaining the SHERLOCK diagnostic kit
In this video, you can learn about the Cas13 nuclease and how it is being used for DNA/RNA detection in SHERLOCK, a CRISPR based diagnostic platform.
Article introducing SHERLOCK (2017)
In this article, you can learn more about SHERLOCK, the CRISPR-based diagnostic tool that is highly sensitive and specific.
Refined version of SHERLOCK announced (2018)
In this article, learn about how the SHERLOCK team developed a simple paper test to demonstrate whether or not a target molecule is present in the sample.
SHERLOCK in Agriculture (2019)
In a recent article, this research team demonstrated SHERLOCK’s use in agriculture by quantifying glyphosate-resistance conferring gene levels from soybeans extracts.
If you prefer other ways to get your CRISPR news, head over to our comprehensive list of all the best CRISPR news sources to find the best option for your favorite way to learn.